FIGURE 2.
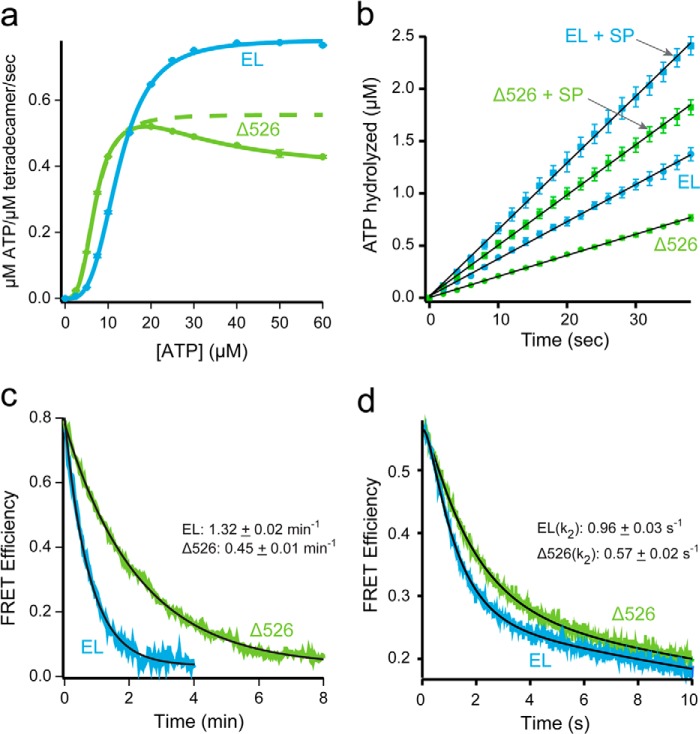
Stimulated folding of Rubisco is not the product of extended GroEL-ES cavity lifetime. a, full-length (EL; blue) or C-terminal deletion (Δ526; green) GroEL (200 nm) was mixed with various concentrations of ATP, and the steady-state rate of hydrolysis was measured. For GroEL, the observed change in initial rate was well fit by the Hill equation, yielding kcat = 0.12 ± <0.01 s−1 per active subunit, K½ = 12.4 ± 0.1 μm, nH = 3.3 ± 0.1. The data for Δ526 at low ATP concentrations was also well fit to a two-state Hill model (green dashed line), yielding kcat = 0.08 ± <0.01 s−1 per active subunit, K½ = 6.9 ± 0.1 μm, nH = 3.2 ± 0.1. The full Δ526 data set was also fit to the nested cooperativity model (52), yielding kcat, 1 = 0.09 ± <0.01 s−1 per active subunit; kcat 2 = 0.05 ± <0.01 s−1 per active subunit, K½ = 1.5 ± 0.1 μm, nH = 3.8 ± 0.1, L1 = 2.5 × 10−3 ± 0.1 × 10−3, L2 = 9.3 × 10−5 ± 2.7 × 10−5. b, steady-state ATP hydrolysis by GroEL (125 nm) and Δ526 (125 nm) was measured in the presence of excess GroES (250 nm), with and without non-native denatured Rubisco (dRub, 100 nm; SP). Addition of dRub to the GroEL-GroES system results in a hydrolysis rate enhancement of 1.8-fold (2.1 μm/min to 3.8 μm/min), consistent with previous observations using saturating levels of the substrate protein malate dehydrogenase (20). Addition of dRub to the Δ526-GroES system results in a rate enhancement of 2.4-fold (1.2 μm/min to 2.9 μm/min). c, lifetime of the GroEL-GroES complex in the absence of substrate protein was examined using a previously described FRET assay (20). ATP (2 mm) was added to GroES-98ED (100 nm) and either 315F-labeled full-length (EL; blue), or C-terminal deletion (Δ526; green) GroEL (125 nm). After a 1 min incubation, unlabeled GroES (2 μm) was introduced as a competitor. The change in FRET was calculated from matched donor-only and donor-acceptor traces. The curves were fit to a single-exponential rate law, yielding rates of 1.32 ± 0.02 min−1 (EL) and 0.45 ± 0.01 min−1 (Δ526). The average of three experiments is shown. d, lifetime of the GroEL-GroES complex upon addition of a stoichiometric amount of non-native Rubisco was examined using the same FRET assay as in c (20). Experiential conditions are similar; except that a stopped-flow apparatus was employed, and 100 nm denatured Rubisco was mixed with the cycling GroEL-GroES system simultaneously with excess, unlabeled GroES. The average of 10 experiments is shown. As reported previously, the observed change in FRET efficiency in the presence of non-native substrate protein requires a triple-exponential rate law for a good fit (20). The rate of the dominant decay component (k2), previously shown to reflect the substrate protein-induced acceleration of GroES release (20), is ∼1 s−1 for GroEL and ∼0.6 s−1 for Δ526.