Figure 2.
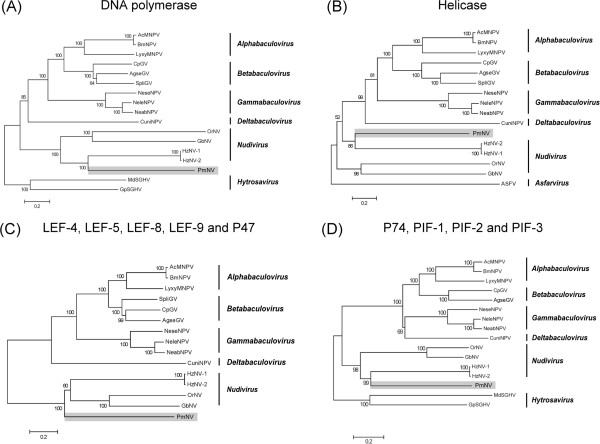
Phylogenetic analysis of selected DNA viruses and PmNV. Multiple alignments of (A) DNA polymerase, (B) Helicase, (C) LEF-4, LEF-5, LEF-8, LEF-9 and P47 (D) P74, PIF-1, PIF-2 and PIF-3 protein sequences were performed using BioEdit. The trees were inferred using MEGA5.2 and the neighbor-joining method. The robustness of each tree was tested using bootstrap (1000) analysis. The percent values are indicated at the nodes. Organisms included in this analysis, with abbreviated names: Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Bombyx mori nucleopolyhedrovirus (BmNPV), Lymantria xylin nucleopolyhedrovirus (LyxyMNPV), Cydia prmonella granulovirus (CpGV), Agrotis segetum granulovirus (AgseGV), Spodoptera litura granulovirus (SpliGV), Neodiprion sertifer nucleopolyhedrovirus (NeseNPV), Neodiprion lecontei nucleopolyhedrovirus (NeleNPV), Neodiprion Abietis nucleopolyhedrovirus (NeabNPV), Culex nigripalpus nucleopolyhedrovirus (CuniNPV), Heliothis zea virus 1 (HzNV-1), Helicoverpa zea nudivirus 2 (HzNV-2), Oryctes rhinoceros nudivirus (OrNV), Gryllus bimaculatus nudivirus (GbNV), Musca domestica salivary gland hypertrophy virus (MdSGHV), Glossina pallidipes salivary gland hypertrophy virus (MdSGHV), African swine fever virus (ASFV). Accession numbers of all the sequences are listed in Additional file 8: Table S6.
