Figure 5.
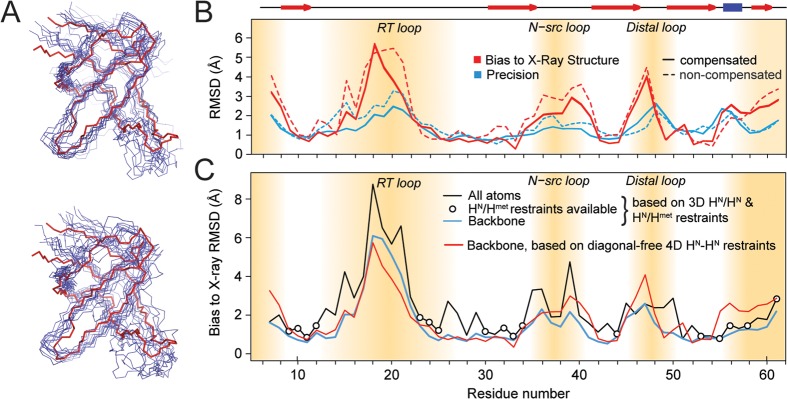
Structure calculation from 1H–1H restraints. (A) Backbone representation of the structural ensembles obtained from diagonal-free 4D correlations superimposed on the reference X-ray structure (PDB: 2NUZ, shown in red). Top: Calculation using peak integrals from diagonal-compensated spectra; bottom: using integrals of the same peaks without compensation. (B) Representation of backbone structure accuracy (bias to the X-ray structure 2NUZ, red) and precision (blue, RMSD to mean) based on high-quality diagonal-free 4D restraints (solid lines). Dashed lines correspond to structures from noncompensated restraints. Secondary structure elements are shown on top with red arrows (strands) and a blue bar (helix). The flexible termini and loop regions are indicated by brown shades, where few contacts are available. All other regions are not far from 1 Å resolution. (C) Comparison of backbone structure accuracy (bias to the X-ray structure 2NUZ) using additional methyl restraints (blue) and using amide–amide contacts only (red). All-atom RMSDs (black) are similar to backbone RMSDs in the presence of amide–methyl restraints (bullets), demonstrating the general potential of methyl-based restraints to further improve accuracy. Methyl data shown here for comparison were acquired using the time-shared experiment in its (truncated) 3D version. See the Supporting Information for details on methyl data.
