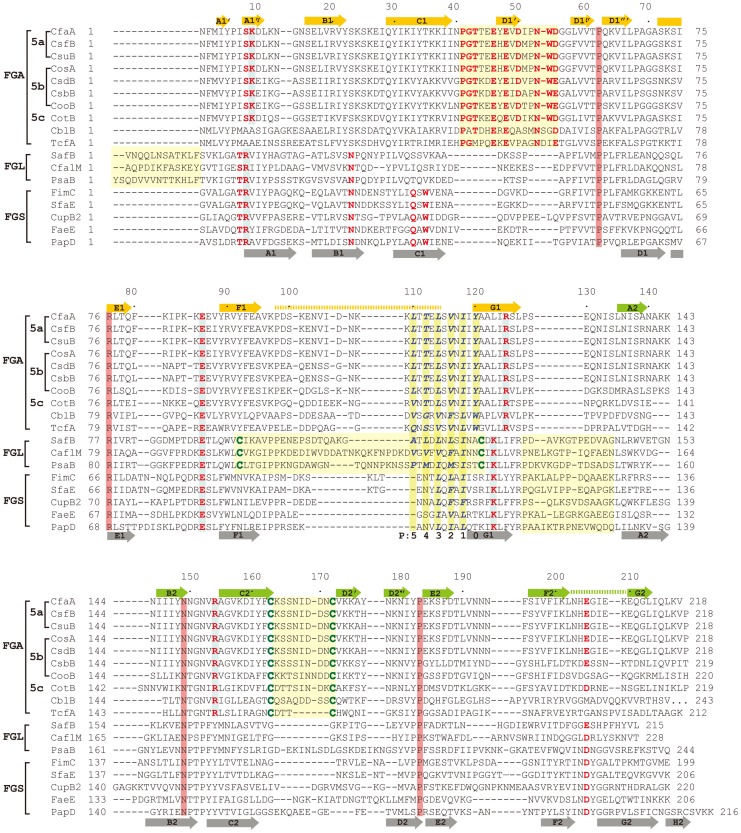
Figure 3. Structure-based sequence alignment of chaperone proteins involved in pilus assembly.
This alignment is based on chaperone structures of CfaA of CFA/I fimbriae (this work), PapD of P pili (PDB:1QPX), FimC of type 1 pili (PDB: 1ZE3), SfaE of S-pili (PDB:1L4I), CupB2 of CupB pili (PDB: 3Q48), FaeE of F4 fimbriae (PDB:3F6L), SafB of Saf pili (PDB:2CO7), and Caf1M of F1 pili (PDB:2OS7). Other included sequences are PsaB of pH 6 fibril of Y. pestis, CsfB, CsuB, CosB, CsdB, CsbB, CooB, and CotB of Class 5 ETEC pili, and chaperones of Class 5 pili from other organisms such as CblA of Cbl pili from B. cenocepacia and TcfA of Tcf pili from S. enterica. The sequence of TcfA has 243 residues and its C-terminal tail is truncated to fit into the figure. The three sub-families of chaperones are referred to as FGA, FGL and FGS. The numbering of secondary structural elements is based on the structure of PapD [2] and are illustrated as gray arrows below the PapD sequence. β-strands for CfaA are shown as orange arrows for the N-terminal domain and green arrows for the C-terminal domain. Dashed orange and green lines indicate disorders in the structure of CfaA. Conserved residues in all chaperones are boxed in red. Conserved residues specific to each subfamily are highlighted in red. Insertions and extensions in each subclass are boxed in yellow. The hydrophobic residues in the alternating pattern of hydrophobic-hydrophilic residues in the donor strand prior to the G1 strand are in italic and boxed in yellow and are indicated at bottom of the alignment as positions P0 to P5. Cysteine residues that form disulfide bonds are highlighted in green and boxed in yellow.