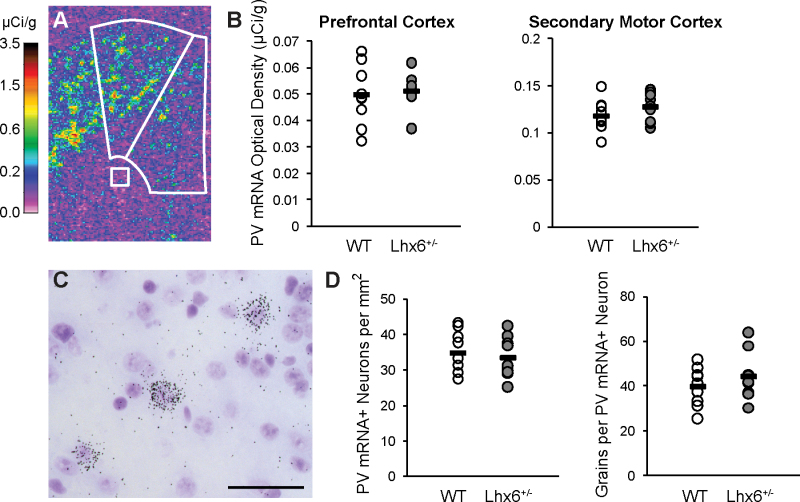
Fig. 3.
In situ hybridization for parvalbumin mRNA in Lhx6+/− mice. (A) Pseudocolored film autoradiograph of a frontal cortex processed by in situ hybridization for parvalbumin (PV) mRNA with calibration scale (µCi/g). White lines denote medial PFC and secondary motor cortex and white matter background measures. (B) Film optical density analysis found that mean parvalbumin mRNA levels did not differ between wild-type (WT) and Lhx6+/− mice in medial PFC or secondary motor cortex. (C) Emulsion-dipped tissue section processed by in situ hybridization for parvalbumin mRNA reveals the presence of silver grain clusters over a minority of lightly Nissl-stained neurons but not over smaller, intensely Nissl-stained glia, which is consistent with specific labeling of parvalbumin neurons. Calibration bar = 50 µm. (D) Cellular grain–counting analyses in the medial PFC found no differences between WT and Lhx6+/− mice in parvalbumin mRNA-labeled neurons/mm2 or mean number of grains per parvalbumin mRNA-labeled neuron.