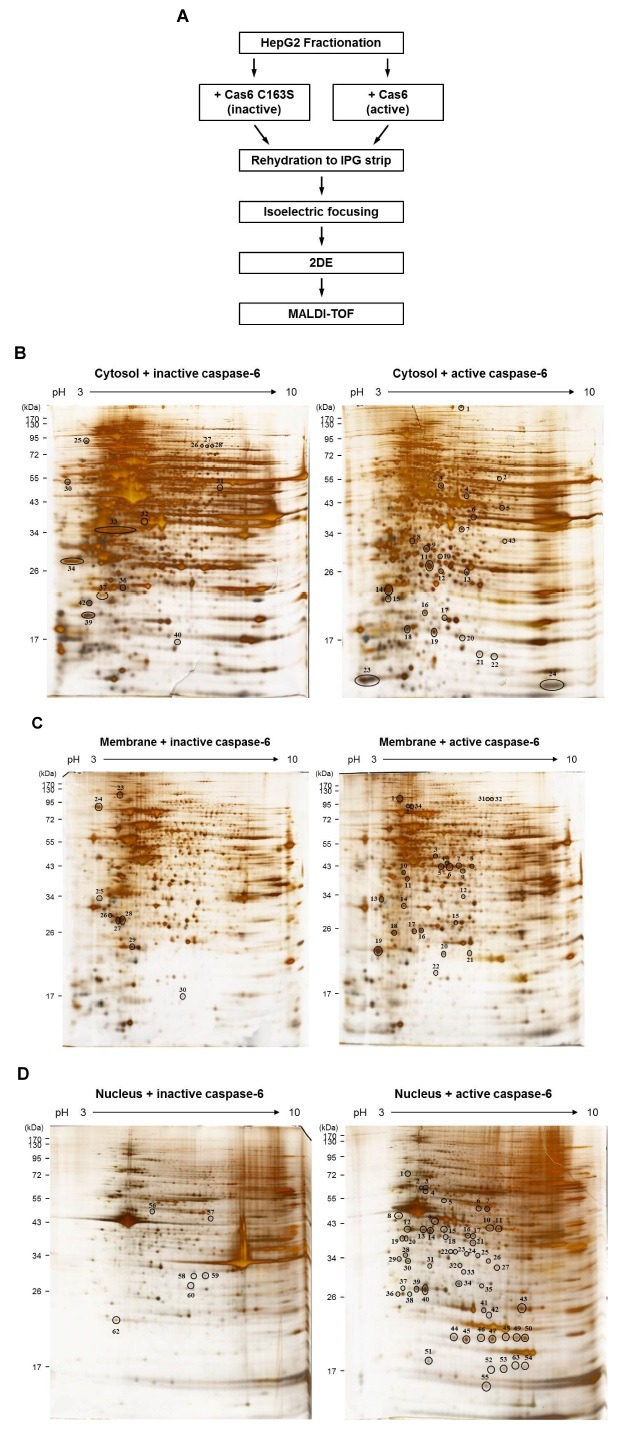
Fig. 1. (A) The workflow for screening to identify novel caspase-6 substrates by proteomics approaches. (B-D) 2-DE gel images showing the differentially displayed spots. (B) Cytosol fractions, (C) membrane fractions, or (D) nucleus fractions, treated with either active caspase-6 or inactive casspase-6, were analyzed using 2-DE. The experiments were repeated for three times, independently. The representative gel images are shown.