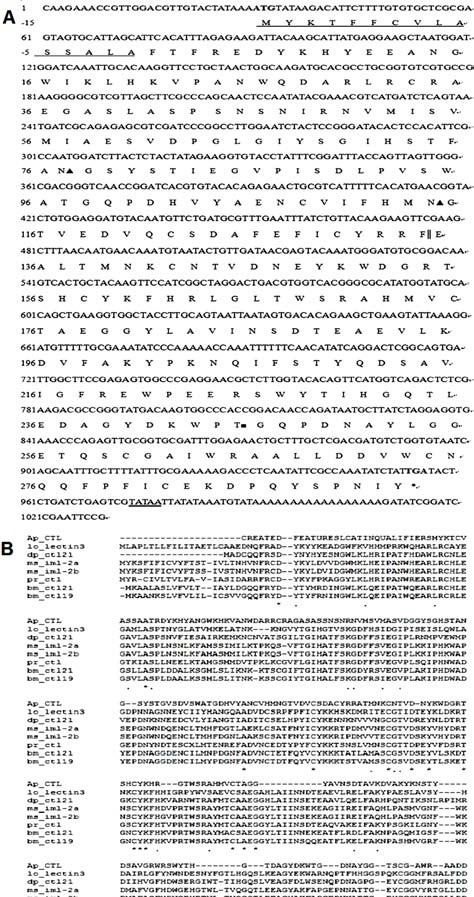
Fig. 1. Sequence analysis and alignment of the cDNA cloning of A. pernyi CTL (Ap-CTL). (A) The numbers of nucleotide and deduced amino acid sequences (one-letter symbols) are shown on the left of the nucleotide sequence, respectively. The predicted signal peptide were assigned negative numbers and underlined. The start codon ATG is shown in bold and the termination cordon (TGA) is marked with an asterisk (*). Two potential N-linked glycosylation sites and one O-linked glycosylation site are marked with ▲ and ■, respectively. The putative polyadenylation sequence, TATAA, is double underlined and the two CRDs are divided by "||". (B) The CLUSTALW multiple sequence alignment program was used to align the A. pernyi CTL amino acid sequence with other 7 most similar proteins. Residues conserved in all sequences are marked with "*", less conservative amino acid subsitutions are marked with ":" and ".", respectively. Ap_CTL, A. pernyi CTL; lo_lectin 3, L. obliqua lectin 3; dp_ctl21, D. plexippus C-type lectin 21; ms_iml-2a, M. sexta immulectin-2a; ms_iml-2b, M. sexta immulectin-2b; pr_ctl, P. rapae C-type lectin; bm_ctl21, B. mori C-type lectin 21 precursor; bm_ctl19, B. mori C-type lectin 19 precursor.