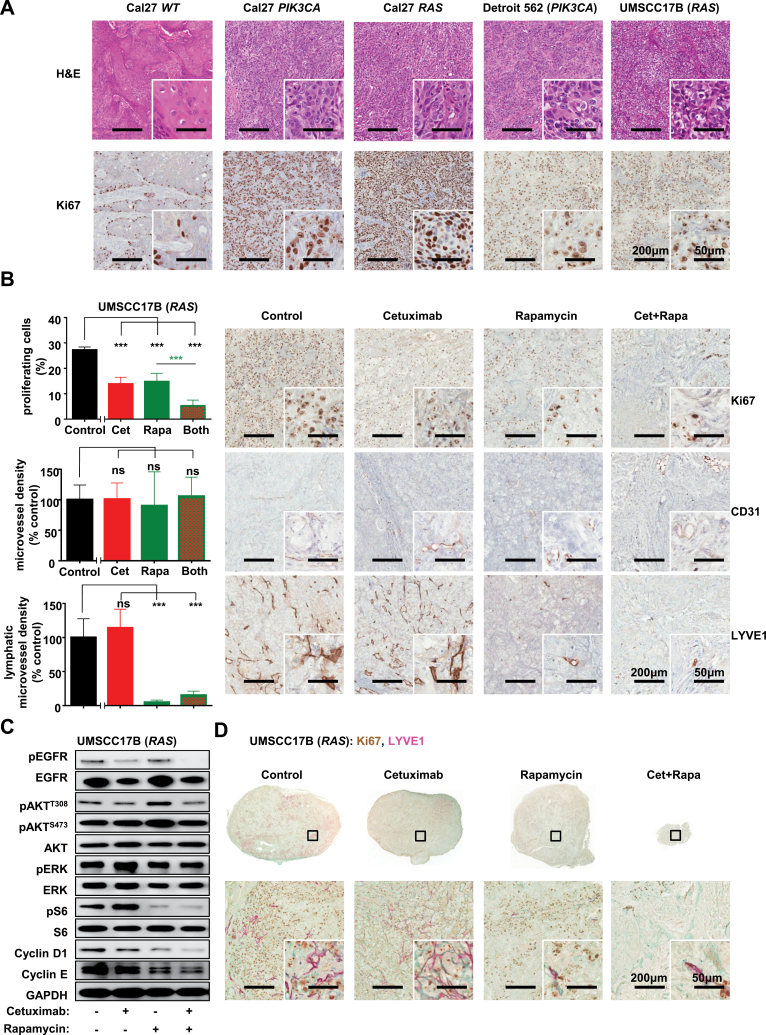
Figure 5.
Effects of the combination of cetuximab and rapamycin on tumor growth driven by PIK3CA and RAS mutations. A) Representative histological sections of tumor xenografts stained by H&E and Ki67 immunohistochemistry as a proliferative marker after transplantation of Cal27 cells, Cal27 cells expressing PIK3CA and RAS oncogenes, as well as Detroit 562 and UMSCC17B HNSCC cells into nude mice. Scale bars represent 200 µm (low magnification) or 50 µm (inset). B) Quantification (left) and representative tumor tissue sections (right) stained for Ki67, CD31, and LYVE1 by immunohistochemistry after a long-term treatment for the indicated days of UMSCC17B xenograft with control, cetuximab, rapamycin, and cetuximab combined with rapamycin, as indicated. (***P < .001 comparing the treatment groups with the control [black stars], or comparing with the indicated treated groups [green stars]; n = 4 per group). The tumor growth curves were compared by the longitudinal data analysis method (two-sided). C) As an example of naturally cetuximab-resistant HNSCC cells, UMSCC17B were transplanted into athymic mice and treated with cetuximab, rapamycin, and with cetuximab combined with rapamycin for four days (short-term treatment). Tumor lysates were analyzed for pEGFR, EGFR, pAKTT308, pAKTS473, AKT, pERK, ERK, pS6, S6, Cylin D1, Cyclin E, and GAPDH. D) Representative tumor tissue sections stained for Ki67 and LYVE1 by immunohistochemistry after a long-term treatment of UMSCC17B xenograft with control, cetuximab, rapamycin, and cetuximab combined with rapamycin, as indicated. Brown color for Ki67, red color for LYVE1.