Abstract
Identifying gene function in specific cells is critical for understanding the processes that make cells unique. Several different methods are available to isolate actively transcribed RNA or actively translated RNA in specific cells at a chosen time point. Cell-specific mRNA isolation can be accomplished by the expression of transgenes in cells of interest, either directly from a specific promoter or using a modular system such as Gal4/UAS or Cre/lox. All of the methods described in this review, namely thiol-labeling of RNA (TU-tagging or RABT), TRAP (translating ribosome affinity purification) and INTACT (isolation of nuclei tagged in specific cell types), allow next generation sequencing, permitting the identification of enriched gene transcripts within the specific cell-type. We describe here the general concept of each method, include examples, evaluate possible problems related to each technique, and suggest the types of questions for which each method is best suited.
Keywords: 4tU-tagging, Gene expression profiling, INTACT, RNA-seq, Transcriptome, Translatome, TRAP.
INTRODUCTION
Identifying gene function within identified cells at a given time point is an important goal for understanding the mechanisms of development. Collecting cell-type-specific transcripts genome-wide (the transcriptome) and transcripts undergoing translation genome-wide (the translatome) in different cell types will help to identify gene networks involved in a process rather than focusing on the effects of single genes one at a time. Transcriptomics refers to a class of high throughput methods that enable measurement of the abundance of tens of thousands of transcribed RNAs (ribonucleic acids) in a given sample. Measuring the relative messenger RNA (mRNA) levels of specific cell types is crucial to understanding the mechanisms that underlie cellular phenotypes. Translatome profiling describes methods that allow the isolation of processed mRNA that is actively being translated in a given sample. Identifying these mRNAs will give the clearest picture of which proteins each specific cell is making at a selected time point. In general, gene expression profiling methods require the isolation and identification of coding mRNAs, which represent only a small portion of a cell’s total RNA pool.
RNA Populations in Eukaryotic Cells
The isolation of mRNAs is complicated by the fact that the majority of RNAs found in eukaryotic cells are non-coding RNAs (ncRNAs). Most are crucial in the process of translation (ribosomal RNA and transfer RNA, accounting for 90-95% of total RNA) while others impact gene regulation (regulatory ncRNAs). Based on their length, regulatory
ncRNAs fall into distinct groups: short ncRNA (e.g. endogenous small interference RNA, microRNA and Piwi-interacting RNA), intermediate ncRNA (e.g. small nucleolar RNA), and long ncRNA [1, 2] (see Table 1). Substantial diversity and lack of strong conservation make it especially difficult to predict lncRNA function [3-5].
Table 1.
Non-coding RNAs (ncRNAs) found in eukaryotic cells.
| Name | Description | Length | Function | Ref. |
|---|---|---|---|---|
| rRNA | ribosomal RNA | 121-5070 nt | organize structure of ribosomal subunits 60S and 30S; crucial for translation | [6] |
| tRNA | transfer RNA | 80 nt | transfer amino acids to a growing polypeptide chain during translation | [6] |
| esiRNA | endogenous small interfering RNA | 21-22 nt | cleave mRNA | [7] |
| miRNA | microRNA | 19-24 nt | prevent protein synthesis through RNA interference; catalyze gene modifications, including methylation, that modify gene transcription activity | [7, 8] |
| piRNA | Piwi-interacting RNA | 26-31 nt | form RNA-protein complexes through interactions with Piwi (P-element induced wimpy testis) proteins; silence transposons | [8, 9] |
| snoRNA | small nucleolar RNA | 60-300 nt | modify RNAs, including methylation and pseudo-uridylation of rRNA and tRNA; can act in regulation of gene expression | [10] |
| lincRNA | long intergenic ncRNAs | > 200 nt | associate with chromatin-modifying complexes | [11] |
| other lncRNA |
long ncRNAs | > 200 nt | regulate protein expression at the level of transcriptional or post-transcriptional processing and chromatin modification; involved in gene silencing; can act as enhancer RNAs | [4, 12] |
Although ncRNAs appear to be important regulators of gene expression, most gene expression profiling studies so far focus on mRNA, likely because ncRNAs constitute a heterogeneous population of different RNAs and many, if not most, ncRNAs are not sufficiently described and their biological relevance is often unknown. Only about 1-5% of all RNAs found in eukaryotic cells are messenger RNAs that code for proteins. Precursor mRNAs (pre-mRNAs) are transcribed in the nucleus and undergo processing to become mature mRNAs, which involves splicing, 5' capping, and 3' polyadenylation. In the cytosol, ribosomes finally translate mRNA to proteins (see Fig. 1).
Fig. (1).
Overview of temporal and spatial RNA isolation methods.
Impact of Different mRNA Populations for Differential Gene Expression Studies
One problem to overcome when analyzing differential gene expression is to choose the population of mRNA within the pool of total mRNA that is optimal for answering the question at hand. Total mRNA consists of newly transcribed mRNA (unprocessed pre-mRNA and newly processed mRNA), steady state RNA (mRNA that was transcribed earlier), actively translated mRNA, and mRNA that is being degraded.
Each population gives insight into distinct aspects of molecular regulation within a cell. Total mRNA analyses, for instance, give a broad overview of the mRNAs contained within cells, comparable with in situ hybridization studies. Acute changes in transcript abundance cannot be measured when analyzing the total pool of mRNAs found in cells. It is important to consider that not all mature transcripts found in a cell are actively translated, but that some are a standing population that is available for translation when needed. Some RNAs have long half-lives, for instance the average estimated half-life in human cells was reported to be 600 minutes [13], or about 420 minutes in mouse embryonic stem cells [14], but many have a half-life of 120-360 minutes and some only 15 minutes [15]. Therefore, a more than 100-fold transient down-regulation in the transcription rate of a single gene would hardly be detectable in total RNA even hours after it occurred without concordant changes in mRNA decay. For the majority of transcripts, a 10-fold up-regulation in transcription rate would require more than two hours to result in a two-fold increase in abundance [16]. When total RNA and newly transcribed RNA were compared, it appeared that many genes are regulated by RNA stabilization and degradation rather than de novo transcription, suggesting that RNA turn-over and synthesis are both important when analyzing differential gene expression levels [17]. The isolation of different mRNA populations, such as newly transcribed or actively translated mRNA requires more sophisticated methods that will be discussed further below (see also Fig. 1), namely 4tU-tagging, translating ribosome affinity purification (TRAP) and isolation of nuclei tagged in specific cell types (INTACT).
One caveat with earlier methods for quantifying gene expression, such as quantitative RT-PCR or microarrays, which were aimed at quantifying expression levels for small or large numbers of genes, respectively, is that they analyze only the standing population of RNA within a cell. These techniques cannot differentiate between actively transcribed, processed or translated RNAs, but rather give an overview of many different RNAs that can be found in cells at a certain time point, including coding transcripts and noncoding RNAs such as microRNAs, small nuclear RNAs and other ncRNAs. One disadvantage of quantitative qPCR is that it is limited to a few genes at a time; a disadvantage of microarrays is that probe sets must be individually designed for each species and analysis is restricted to genes that happen to be on the chip, meaning that non-annotated genes are generally not included and will therefore not be detected, and that species with unannotated genomes cannot be studied genome-wide. Currently, generally commercially available probe sets for vertebrates are available including human, mouse, zebrafish and chicken, but are not available for species of interest such as stickleback or sea urchin.
Next generation sequencing (RNA-seq) can identify transcripts from large numbers of genes over a great dynamic range, not restricted to known genes, not limited to annotated genes, nor restricted to model organisms [18]. Despite these huge advantages, RNA-seq performed on RNA isolated from regular tissue dissection, dissociation, and cell sorting reveals only steady-state levels of mRNA. This includes not only newly synthesized mRNA, but in addition, mRNA made hours before depending on the turnover and half-life of each individual transcript. Furthermore, transcriptional and posttranscriptional regulation, for example, by microRNAs, can influence steady-state mRNA populations and can alter the picture of transcript quantity and diversity found at specific time points.
Sampling Methods to Achieve Regional Selectivity
Even with the technical possibility to retrieve millions of reads that correspond to thousands of RNAs, measuring changes in gene expression from rare cell types is challenging because they contribute only a small fraction of the total tissue RNA. Many tissue-specific transcriptome analyses rely on time-consuming dissection techniques, including manually collected tissues or more sophisticated procedures such as laser capture microdissection or flow cytometry. Given the limited amount of RNA contained in a single cell, which is about 10 picograms total RNA or on average only 1 picogram mRNA, which is equivalent to a few hundred thousand molecules transcribed from about ten thousand genes in humans [19], single cell methods are generally more prone to producing false negatives, particularly for low abundance transcripts [20]. Further, manually collected tissue can lead to variability because of dissecting irregularities resulting in contamination of non-target cell or tissue types. Mistakes in dissection can immensely bias results, especially if only few cells within a complicated tissue type are desired. Because small sample sizes could affect the analysis by under-representation of rare transcripts, pooling cells from the same cell type and analyzing their transcriptome would likely better represent gene expression levels within the target cell type.
Some tissues are not suitable for mechanical dissections, including laser dissection, for example hard tissues such as bone or cartilage. Further, cells containing fat or collagen generally provide poor quality cryostat sections for laser dissections and delicate cells such as mammalian ovarian epithelium and distributed cells like interrenal cells are generally unsuitable for dissection. Other dissociation methods, such as fluorescence-activated cell sorting (FACS), are precise but require chemical treatments that can cause additional cell stress leading to unauthentic gene expression levels [21].
To ensure cell-type specific gene expression, new methods have recently been developed to guarantee reliable isolation of RNA from selected cell types in sufficient quantities. These new technologies depend on various methods for labeling RNA before isolation. In addition, these methods can add temporal control by isolating either actively transcribed or translated genes at different stages of development, in contrast to analyzing only the steady-state levels of RNA within a cell; thus, these methods allow the identification of genes that are differentially expressed over time on a genome-wide scale.
Several methods developed to isolate RNA populations are shown in (Table 2). Among those methods, the following are especially suitable for isolating RNA with spatial and temporal resolution (see Fig. 1): biosynthetic labeling of RNA (TU-tagging [22-24], also known as RNA Analysis by Biosynthetic Tagging or RABT [25]), and ribosomal tagging methods (translating ribosome affinity purification or TRAP [16], also known as RiboTag [26]) allow isolation of biologically relevant transcripts such as actively transcribed RNA or actively translated RNA, respectively, at any chosen time. Another approach to isolate temporally relevant RNA is to isolate nuclei tagged in specific cell types (INTACT) [27]. Below, we examine these recent techniques in more detail.
Table 2.
Overview of methods used to isolate RNA from specific cells.
| Method | Protocol | Output | Caveat | Ref. |
|---|---|---|---|---|
| 4tU-tagging | Uprt enzyme transforms 4-thiouracil to thio-UMP, which can be incorporated into RNA, followed by biotin-streptavidin purification and RNA-seq | Biosynthetically labeled newly transcribed RNA | Contamination by nonspecific transcripts, ribosomal RNA depletion required before purification | [28] |
| TRAP | N-terminal fusion of GFP to RpL10 allows polyribosome immunoprecipitation using GFP antibodies, followed by mRNA isolation and RNA-seq | Actively translated ribosome-associated mRNAs | Contamination by nonspecific transcripts; doesn’t isolate non-coding RNAs | [23, 29-31] |
| Ribo-Tag | RpL22-3xHA fusion protein allows polyribosome immunoprecipitation with HA antibodies and mRNA purification | Actively translated ribosome-associated mRNAs | Contamination by nonspecific transcripts; doesn’t isolate non-coding RNAs | [26, 32] |
| INTACT | Nuclear targeted fusion protein allows biotin-streptavidin purification, followed by nucRNA-seq and ChIP-seq | Unspliced primary transcripts and nuclear-retained long non-coding RNAs | Assumes that nuclear RNA correlates with primary transcript frequency | [27, 33, 34] |
| Laser capture or laser directed microdissection (LCM and LDM) | Uses laser to isolate and capture cells | Steady-state mRNA and other RNAs | Contamination by nonspecific transcripts, small sample sizes; some cell types unsuitable for LCM; cells are fixed or frozen | [35] |
| FACS (Fluorescence-activated cell sorting) | Separates dissociated cells by fluorescence or scatter, and sorts cells into receptacles | Steady-state mRNA and other RNAs | Could induce stress response during isolation | [36] |
| PAN (“Panning”) | Unlabeled dissociated cells purified using cell type specific antibodies using panning plates | Steady-state mRNA and other RNAs | Requires specific antibody, time consuming; dissociation, delays, and reagents could induce stress | [37] |
BIOSYNTHETIC LABELING OF NEWLY TRANSCRIBED RNA
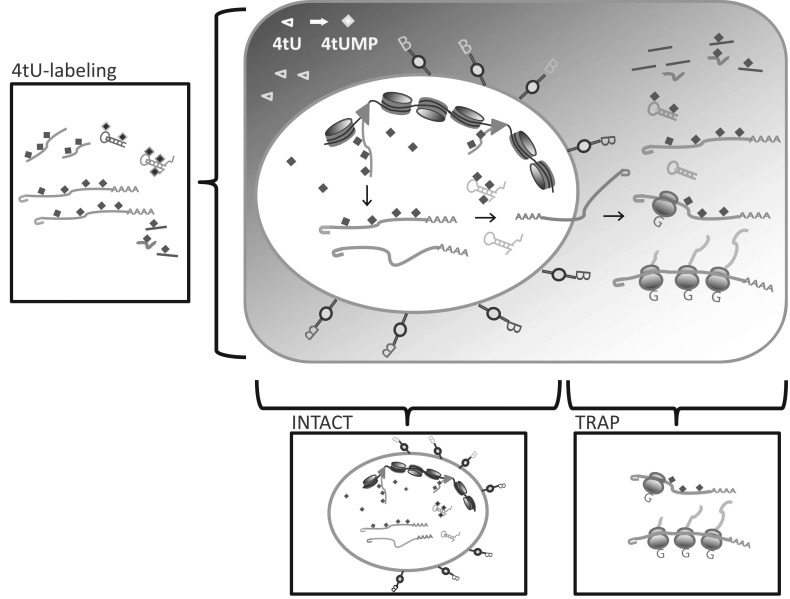
Labeling RNA with thiol groups provides a tool to enrich for newly made mRNA in extracts from tissues or from whole animals [28] (Fig. 2). Thiol-labeled nucleotides are not a natural component of nucleic acids, but can easily diffuse into cells from the media or bloodstream. Thiol groups are incorporated into newly made RNA whenever the substrate is available, thus giving the researcher temporal control of thiol-labeling. Two RNA precursors have been used for RNA thiol-labeling: the nucleotide thiouridine (4sU) and the base thiouracil (4tU). Thiol-labeled RNAs can be purified readily from other RNAs and quantified accordingly.
Fig. (2).
Schematic overview of the method 4tU-labeling.
Purification of Thiol-labeled RNA
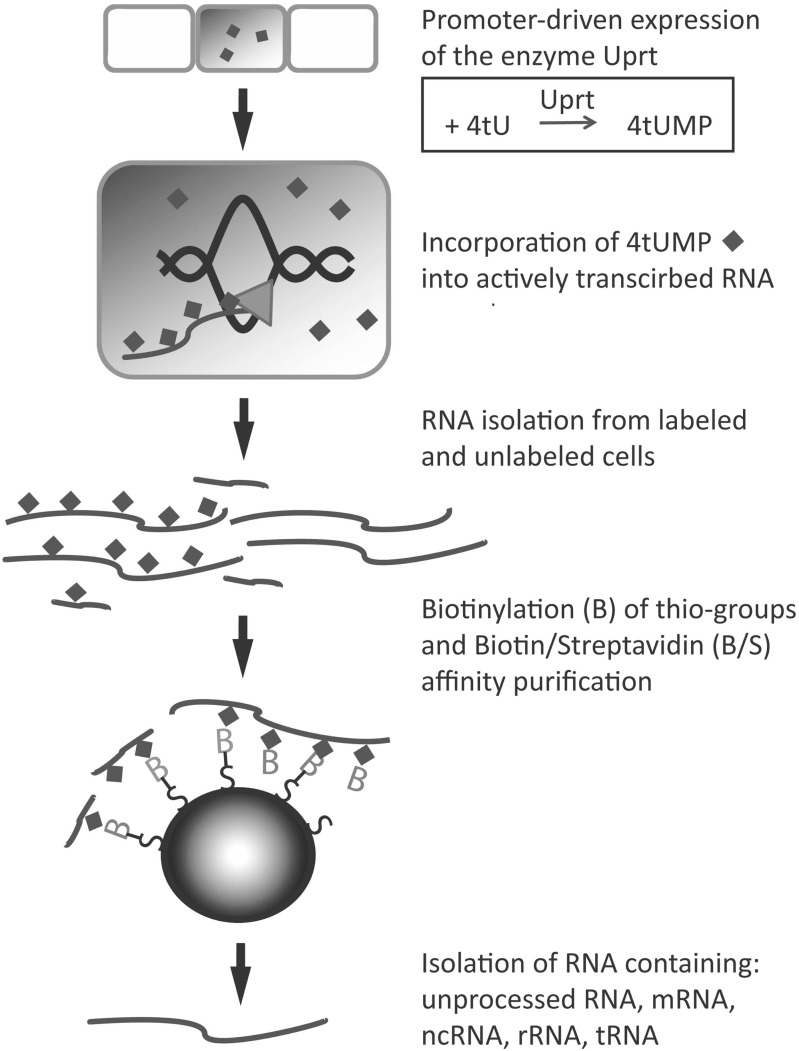
Thiol-labeled RNA can be quantitatively biotinylated using thiol-reactive reagents. One reagent commonly used is biotin-HPDF (N-[6-(Biotinamido)hexyl]-3'-(2'-pyridyldithio) -propionamide), a pyridyldithiol-biotin compound that labels molecules that contain sulfhydryl groups, including cysteines in proteins. This reaction creates a reversible disulfide bond linking biotin directly to the thiol-containing nucleotide. The thiol-labeled RNA-Biotin complex can then be detected using Streptavidin-HRP or purified using Streptavidin-coated magnetic beads [22]. Purified RNA can be further processed and analyzed by qPCR, microarrays, or RNA-seq. Thiol-labeling is not restricted to newly made mRNA, but is also incorporated into newly made ncRNAs and rRNAs. Because rRNAs represent a large portion of total RNA within a cell, rRNA must be eliminated from the sample before further purification or sequencing.
4sU-labeling of Newly Made RNA
By taking advantage of the pyrimidine salvage pathway [38], most eukaryotes can use the nucleotide 4sU as a substrate for RNA synthesis [39] and the 4sU can become incorporated selectively into newly-made transcripts [40]. This provides an advantage of 4sU over 4tU because no modifications are required prior to the experiment, such as creating transgenic lines. Because all cells in an animal will make thiol-labeled mRNA when the thiol-labeled 4sU is available, regional selectivity must be accomplished by physical manipulations, such as dissection or FACS. 4sU-labeling in cell culture and in intact animals demonstrated enrichment of up to 4-fold, validated by microarray, and about 7-fold, validated by qPCR, of tissue-specific genes in dissected kidneys of 4sU-injected mice [23].
4tU-labeling of Newly Made RNAs is Restricted to Specific Cells
A more sophisticated method to enrich for newly made mRNAs is biosynthetic tagging of RNA in a tissue-specific manner using 4tU. Similar to 4sU, the use of 4tU as substrate isolates actively transcribed genes expressed in specific cells only during the period in which the substrate is available to the cells. In contrast to 4sU, which is incorporated into every cell exposed to the compound, the use of 4tU allows cell-specific labeling. Because animal cells can not readily produce thiol-labeled uridine from thiol-labeled uracil, the production of transgenic animals expressing a protist enzyme that converts thiouracil to thiouridine allows labeling of newly made RNA in specific cell types.
Cells of interest are defined by a promoter driving the expression of the pyrimidine salvage enzyme uracil phosphoribosyl-transferase (Uprt) from the parasitic protist Toxoplasma gondii [22, 28, 41]. Uprt makes 4-thiouridine monophosphate (4tUMP) by coupling ribose-5-phosphate to the N1 nitrogen of 4tU; the resulting thiol-labeled nucleotide can be incorporated into newly made RNA (for details see Iltzsch, 2007 [42]). Comparisons of the T. gondii Uprt sequence to Uprt-like proteins in other organisms identify orthologs with varying degrees of sequence similarity. In addition to T. gondii, only prokaryotes (archaebacteria and eubacteria) and unicellular eukaryotes such as yeast cells can convert 4tU to 4tUMP by their endogenous Uprt enzyme. Proteins containing a Uprt domain have been reported in higher multicellular eukaryotes, including humans. However, modifications in the domain associated with uracil binding suggest that higher eukaryote orthologs diverge in function from T. gondii Uprt [43]. Indeed, assays in human and mouse cells have shown that Uprt orthologs are unable to convert uracil to UMP [22]. In vertebrate or fly model systems, only transgenic cells that express Uprt under control of the promoter of interest can use 4tU as a substrate to synthesize RNA; labeled RNA can then be purified from total RNA (Fig. 2) as described in more detail above.
Another possibility for driving the expression of the enzyme Uprt in specific cells is the use of the modular expression system Gal4/UAS [44]. Any line expressing Gal4 in a tissue-specific fashion can be used together with an UAS:uprt effector line to express Uprt in the tissue of choice. This option is limited to available Gal4 lines but more and more lines are available in different animal model systems such as fly [45], zebrafish [46, 47] and mouse [48] and can be readily constructed. Another option is using the Cre-lox system [49], which has been used successfully in mouse to express Uprt in specific tissues [50].
The 4tU-tagging method is useful for a variety of experimental protocols. For example, pulse-chase experiments with 4tU can demonstrate changes in transcript abundance and transcript decay [16] over time. Furthermore, transcriptome profiling assays using 4tU-labeling followed by microarray and RNA-seq analyses show that this technique can be successfully adopted in cell culture [23], in Drosophila [24] and in mouse [50]. Researchers can introduce 4tU into cells by different methods depending on the animal model. Like 4sU, 4tU can diffuse through the organism, even crossing the placenta and the blood-brain barrier [50]. 4tU can be fed to adult flies, and fly embryos or cell cultures can be incubated in media containing 4tU [22, 24, 51]. Mouse embryos can be exposed by injection of 4tU into the mother [50]. Zebrafish (Danio rerio) embryos can be incubated in 4tU solution (unpublished observation A.T., J.H.P, P.E.W). Incubation time and 4tU concentration vary depending on the strategy used and must be optimized to achieve the best results [28]. Under optimized conditions, differences between biological replicates have been described to be small and results have been highly reproducible [24, 50, 51].
Kinetics of Thiol-group Incorporation
To understand the enrichment of thiol-labeled transcripts, it is important to consider the kinetics of thiol-group incorporation. Shorter transcripts or transcripts with fewer U nucleotides would incorporate fewer thiol-groups and might be underrepresented or lost from the analysis. The question of thiol-group incorporation has been addressed using 4sU-labeling in cell culture. The approximate rate of 4sU incorporation into total RNA was calculated to be four to five 4sU molecules/1 kb of RNA [23]. The thiol-content can be directly quantified by spectrophotometric measurements by absorption of light at 330 nm [52]. The density or number of 4sU molecules necessary for purification of an individual transcript molecule has not yet been determined. Assay sensitivity also depends on the efficiency of biotin labeling of the thiol-groups. Given these considerations, careful thought is needed to design appropriate controls.
Kinetics of mRNA Processing
Knowing the time-line of different mRNA processing events is critical for finding the optimum method to isolate newly made mRNAs from pools of already processed and currently translated mRNAs. This consideration is especially important because thiol-groups are incorporated into both exons and introns of newly synthesized mRNA. The kinetics of RNA transcript processing and regulation remain elusive. In an attempt to unravel the kinetics of intron processing, Windhager et al. used 4sU, which is only incorporated into newly synthesized RNA, in human B-cells [17]. This method enabled the group to focus on the presence of introns in newly made transcripts by applying 4sU for periods of time ranging from 5 to 60 minutes. Results showed that many introns had already been removed by 5 minutes, confirming other reports describing rapid degradation of introns after correct splicing [16, 52, 53]. Interestingly, splice rates showed large variability, with a large number of introns being retained when incubation times shorter than 60 minutes were used [17].
This finding leads to the discussion of whether pre-mRNA transcripts that still contain introns should be removed before analyzing transcriptomic data to reduce the risks of including data from genomic DNA contamination. In general, intron retention and slow splicing processes are probably rather uncommon and therefore introns are most commonly excluded from transcriptomic analyses. To sample the level of genomic DNA contamination, one could compare the number of reads in the sense direction of the intron with that in the antisense direction. The antisense would be genomic DNA, the sense would include the same amount of genomic DNA contamination plus the bone fide intronic transcript. However, sense-antisense transcript pairs that are involved in post-transcriptional regulation of gene expression would also fall into the genomic DNA contamination category (15% of mouse genes and 8.4% of human transcription clusters form predicted sense-antisense pairs) [54]. Another option to eliminate genomic contamination is to use different RNA clean-up methods, for instance only isolating mRNAs using commercially available mRNA purification systems.
Limitations of 4tU-labeling
In practice, 4tU-labeling has some limitations to consider. Although, in theory, only Uprt-expressing cells should be able to incorporate 4tU, several reports have mentioned detectable background signals that compromise the sensitivity of this technique [24, 28, 50]. Background noise could originate from unlabeled DNA and RNA that is pulled down together with the 4tU-labeled RNA during the thiol-biotin purification step. Further, limited 4tU-labeling could occur in cells not expressing the Uprt transgene, discussed in more detail below. In principle, isolation of thiol-tagged RNA should be possible without any mechanical dissection, but in practice, the sensitivity of this method is limited by the ratio of the amount of labeled RNA from cells expressing the Uprt transgene to the amount of label that arises from cells not expressing the transgene. Therefore, the percentage of cells expressing Uprt in the whole sample appears to be critical for the success of the experiment [24, 50]. So far, successful results were obtained when about 1-5% of cells in the extracted sample carried the transgene [24, 50]. Increasing the percentage of cells expressing the Uprt transgene can be achieved by dissecting out specific organs. For example, when working with a few neuronal cells that express uprt in the developing fly brain, heads had to be dissected from the body [24]. Using the entire organism led to high background levels in Northern blot analysis, showing almost similar signal levels of total RNA compared to 4tU-tagged RNA. Using a minimal dissection (i.e. using whole heads as a ‘brain’ dissection) showed that thiol-labeled RNA was specifically incorporated into Uprt-expressing cell types, as shown by microarray analysis.
Efficiency of 4tU-labeling in Mouse
Experiments with different tissues give insight into the efficiency of 4tU-labeling under various experimental conditions. In mouse, endothelial cell RNA in both heart and brain [50]. While Uprt expression in the heart was abundant, with about 70% of cells expressing Uprt, expression of Uprt in endothelial cells in the brain was more restricted, with only 5% of brain cells expressing Uprt. RNA-seq analysis clearly demonstrated enrichment of tissue-specific transcripts in the brain and in the heart. While the vast majority of genes were expressed roughly equally in both endothelial and neural cell types, 11 of 13 previously identified pan-endothelial control genes showed an average of 3.9-fold enrichment in the 4tU-tagged RNA from brain samples. 76% of 130 enriched genes showed highly specific expression in endothelial cells or microglia and macrophages in the brain and only 1% of enriched genes were expressed broadly in neurons, suggesting that there is only low background contamination from transcripts that are expressed in cells not containing the Uprt transgene. In 4tU-tagged cells from developing mouse embryos, less than 50% of control genes known to be expressed in endothelial cells were detected, and only 33% of the enriched genes appeared to be specific to the expected cell types, based on the Eurexpress database, a transcriptome atlas database for the mouse embryo. Differences in results obtained from postnatal versus embryonic stages were hypothesized to result from either less 4tU reaching the embryos or lower expression of endothelial genes in the embryonic brain. The authors did not further test these possibilities, for instance by quantifying the amount of 4tU in embryonic or postnatal mouse brain or qPCR studies of endothelial gene expression in embryonic versus postnatal brain. In the heart, 72% of enriched genes were found to be specifically associated with endothelial and endocardial cells or Tie2:Cre lineage-derived atrio-ventricular canal cushion mesenchymal cells.
In summary, to obtain maximum enrichment and statistical significance, the ratio of 4tU labeled RNA to total unpurified RNA must lie within certain ranges. If cells expressing Uprt are too abundant, differences from total RNA will be too small. On the other hand, if only a few cells express Uprt and, assuming a short 4tU pulse (<1h), only about 10% of the newly synthesized population of transcripts in the Uprt-expressing cells will be 4tU-labeled [22], then the number of transcripts might be insufficient to show statistical enrichment compared to total RNA. The number of Uprt-expressing versus non-expressing cells should lie at least above 1% [50].
Possible Sources for High Background
Any of the several sources mentioned below could contribute to high background levels in 4tU experiments. Unexpectedly high background levels from tissues might be explained by contamination of the tissue sample with bacteria that can also incorporate 4tU in their RNA [51]. Also unlabeled RNA could contaminate the purified thiol-labeled RNA fraction due to nonspecific binding of RNA to the magnetic beads or by overloading the beads.
Another possibility is that 4tU might label RNA in cells that don’t themselves express Uprt. In mammals, biochemical pathways that do not require Uprt can also convert uracil to UMP, but they are typically much less efficient than Uprt. In mammals, enzymes that could sequentially convert 4tU to thiol-labeled 4tUMP include orotate phosphoribosyltransferase (the penultimate enzyme in the de novo pyrimidine synthesis pathway), pyrimidine phosphorylase and uridine kinase [51]. These reactions should occur at a low rate compared to the Uprt reaction [55-57] but the activity of these enzymes have not been directly compared to the T. gondii Uprt enzyme. Charged nucleosides such as UMP, and probably 4tUMP, cannot easily pass through the cell membrane and rely on specialized carrier proteins [58, 59] for their membrane translocation which might allow cells not expressing Uprt to make 4tU-labeled transcript. Although these scenarios together should account for only a fraction of nonspecifically 4tU-labeled RNA, additional experiments are necessary to clarify unresolved questions about 4tUMP synthesis in cells not expressing Uprt and to determine the selectivity of 4tUMP to permeate membranes. Leakage of 4tUMP into cells not expressing Uprt might interfere with analysis, in particular in tissues where only a small percentage of cells express Uprt or where long labeling times are applied.
concluding Remarks to Thiol-labeling
4tU-tagging allows cell-specific labeling and collecting of newly made transcripts during a controlled time period by incorporating thiouracil into newly made RNA, as well as coding and non-coding RNAs. One disadvantage of 4tU-tagging is that certain parameters such as the percentage of cells expressing Uprt have to be maintained in a specific range to obtain significant differences of sample over background levels. A strong advantage of 4tU-tagging is that it allows the purification of noncoding RNAs in addition to coding RNAs, which current data sets mostly focus on. In the future, it would be of great interest to include miRNAs and other functional ncRNAs in the analysis, with the caveat that recovery might be low for short transcripts containing few uracils.
TRANSLATING RIBOSOME AFFINITY PURIFICATION (TRAP)
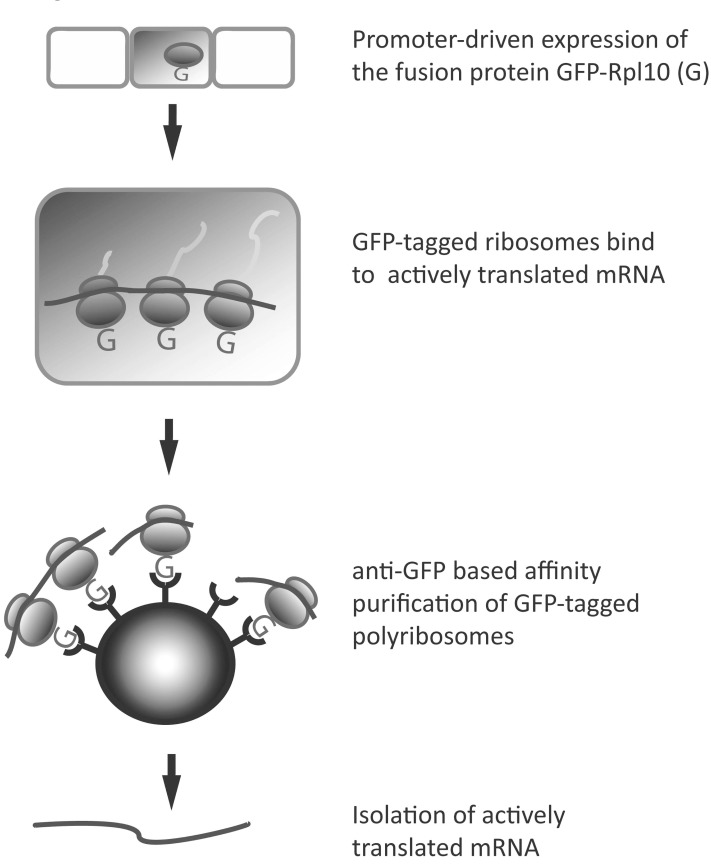
While 4tU-labeling identifies newly made RNAs, it does not reveal which mRNAs are being actively translated; in contrast, TRAP allows quantification of translated mRNAs. TRAP (Fig. 3) isolates transcripts by GFP-mediated polysome immunoprecipitation directly from tissue homogenates [30]. This system requires a selected promoter, or the UAS/Gal4 or Cre-lox system, that drives the cell-type specific expression of a fusion protein consisting of GFP and the large ribosomal subunit L10 (rpL10). Ribosome-associated mRNA can be affinity purified using bead-conjugated anti-GFP antibodies that specifically bind to GFP-RpL10 protein, most of which should be located in polysomes [30]. If the desired transgenic animals are available, then no further chemicals or substrates need to be applied to allow isolation of cell-specific transcripts.
Fig. (3).
Schematic overview of the method TRAP.
By isolating ribosome-associated mRNAs, TRAP identifies mRNAs that are being actively translated, giving insight into which proteins are crucial for cell function at that time. Given the “snap-shot” nature of this technique, mRNA transcripts that are in the process of being transcribed, but are not yet associated with ribosomes will not be taken into account. The origins of TRAP reside in early translational profiling techniques, such as polysome (or polyribosome) fractionation [60], improving the technique by adding cell specificity.
Limitations of Polysome Purifications
Using the TRAP method allows the prediction of protein abundance within specific cells. Investigating which mRNAs are actively translated helps to understand protein expression during a specific time period in specific cells. However, genes with the most abundant transcripts within a certain cell type are not necessarily those that are most active in translation and hence that are present at the highest level in the ribosome [61, 62]. It is important to consider that protein abundance also depends on rate of translation and protein degradation rates [63]. Translation elongation speed is a critical regulator of translation efficiency and therefore protein synthesis [64] and varies among genes and along the transcript sequence depending on the coding sequence in its vicinity, concentrations of elongation factors and tRNA molecules and mRNA folding strength. Further, TRAP cannot measure the activity of proteins as some proteins require posttranslational modifications such as phosphorylation or ubiquitinylation or structural changes such as proteolytic cleavage in order to be active.
More experiments are necessary to determine the exact time frame in which translatome profiling can efficiently measure changes in transcript translation, depending on factors such as GFP-tagged ribosome degradation or GFP-tagged ribosome synthesis and ribosome binding to the mRNA. Not just ribosome occupancy but, more importantly, ribosome density is the critical factor for efficient transcript isolation. Further, the length of individual transcripts might be an important factor for efficient pull-down and one could assume that longer mRNAs are associated with more ribosomes and so are more likely to be immunoisolated, even though the density of ribosomes on the message might be the same as a short mRNA. However, Doyle et al. did not find a bias towards longer transcripts when plotting signal intensity versus transcript length [29]. This result might be explained by the observation that shorter transcripts often show higher ribosome densities than longer transcripts, suggesting an inverse correlation between mRNA length and ribosome density [65]. Therefore, ribosome density is highly variable and would need to be determined for every single transcript.
One characteristic of the TRAP method is that the full population of already translated RNAs, non-coding RNAs and unprocessed RNAs that might be important for gene regulation will not be accounted for using this method (see Table 3). Stress response experiments in yeast showed responses of gene expression levels to induced severe stress already after 10 minutes, monitored by polysome profiling [66]. However, some ncRNAs that can be associated with ribosomes [67], such as long intergenic ncRNAs (lincRNAs), will be isolated by this method together with translated coding mRNAs [68]. It should be possible to remove lincRNAs bioinformatically from the data set as they differ in some aspects from mRNAs. The pattern of ribosome occupancy on the lincRNAs is comparable to other known noncoding RNAs or untranslated regions (UTRs) of transcripts; for instance, most lincRNAs do not show the sharp decrease in ribosome occupancy following a stop codon that is seen for known protein-coding RNAs [67].
Table 3.
Comparison of characteristics for 4tU-labeling, TRAP and INTACT methods.
| 4tU-labeling, pulse (< 1 hour) |
4tU-labeling (several hours) |
TRAP | INTACT | |
|---|---|---|---|---|
| mRNA | + | + | + | + |
| rRNA | + | + | + | + |
| tRNA | + | + | - | + |
| non-coding RNA | + | + | + (subset) | + |
| unprocessed RNA | + | + | - | + |
| highly dynamic RNA populations | ++ | + | + | + |
| “steady-state” RNA pool | - | + | + | +/- |
| cell-type specific RNA population | + | + | + | + |
| identifies differences in RNA expression before and after treatment or in disease models | ++ | + | + | + |
| possible biases | rRNA is highly abundant and could bias RNAseq libraries, especially if the number of mRNA transcripts is limited | rRNA abundance | some long intergenic noncoding RNAs display low ribosome binding; differences in ribosomal occupancy of transcripts | rRNA abundance, transcripts encoding proteins regulating transcription and mitosis might be overrepresented |
| purification steps described before RNAseq | rRNA depletion | rRNA depletion | - | none described |
Combining translatome profiling with transcriptomics, such as 4tU-tagging, and comparing actively transcribed with actively translated mRNAs will help in understanding the big picture of gene regulation and protein synthesis within specific cells or tissues.
Efficiency of TRAP Analysis in Mouse
We know that TRAP analysis has the potential to accurately identify enrichment in specific cell populations because Doyle et al. found known markers for each cell type in their datasets [29]. In a transgenic mouse approach, called BACarray translational profiling, the authors generated numerous Bacterial Artificial Chromosome (BAC) lines driving the expression of the EGFP-tagged ribosomal protein (EGFP-RpL10a), that allow TRAP analyses of protein expression levels in many different neuronal tissues [29, 30]. Biological replicates for the same cell type showed little variance for genome-wide translational profiles (average Pearson’s correlation was above. 0.98). In situ hybridization verified gene expression of TRAP-identified but formerly unknown transcripts within specific cell types. The average marker gene enrichment across cell types showed a 2 to16-fold enrichment of BACarray RNA to whole tissue. Tissues with a low number of transgene expressing cells or conversely tissues in which almost all cells express the transgene EGFP-RpL10a, failed to show significant enrichment of tissue-specific genes. These results suggest that the ratio of transgene-expressing cells to non-expressing cells needs to be within certain limits to guarantee values that allow differential gene expression analysis. qPCR of some marker genes confirmed enrichment in TRAP-isolated RNA compared to total RNA (about 2 to 60-fold change). Interestingly, when the authors looked for functional categories that were significantly over-represented in the nervous system, they found more transcripts encoding cell surface proteins (channels and receptors) than transcription factors and calcium binding proteins. The authors suggest that neuronal diversity is primarily driven by the expression of cell surface proteins and to a lesser extent transcription factors and calcium binding proteins. An alternative explanation is that some transcripts might be more accessible for TRAP purification, based on their location of translation (ribosomes bound to the endoplasmic reticulum or free ribosomes in the cytosol).
A recent study by Zhou et al. reported the generation of a mouse line Rosa26fsTRAP allowing Cre-activated expression of GFP-RpL10 [68]. The authors analyzed Cre-mediated specific expression of GFP-RpL10 in endothelial cells driven by Tie2-Cre, and in cardiomyocytes driven by cardiac troponin T (TNT)-Cre. They reported that in heart tissue, endothelial marker genes were 10-fold enriched in Tie-TRAP samples and less than approximately 0.2-fold enriched in TNT-TRAP samples, whereas cardiomyocyte-specific transcripts were depleted in the endothelial Tie-TRAP sample. The cardiomyocyte TNT-TRAP sample, however, did not show significant enrichment of cardiomyocyte-specific transcripts, likely due to the abundance of cardiomyocytes within the heart, resulting in only minimal differences in levels of TRAP-isolated transcripts and total cytoplasmic transcripts. Evaluation of RNA-seq data from heart samples suggested differential expression of more than 1500 genes between endothelial Tie-TRAP and cardiomyocyte-specific TNT-TRAP samples. About 908 genes showed endothelial enrichment levels with the ratio of Tie2-TRAP RNA to total cytoplasmic RNA greater than five, and strong endothelial expression with more than 10 reads per kilobase transcript per million reads (RPKM). Further, the authors analyzed TNT-TRAP RNA-seq data in an introduced heart disease model, namely aortic banding or sham operation, finding differentially expressed genes in isolated TRAP RNA or total cytosolic RNA in both conditions. The authors confirmed the correlation between fold change measured by RNA-seq and qPCR for some of the genes suggesting that TRAP is suitable to identify genes that are differentially expressed in cardiomyocytes in a disease model.
TRAP in Other Model Systems: Drosophila, C. elegans and zebrafish
In Drosophila, Thomas et al. [69] showed that small cell populations can be profiled using TRAP. The fusion protein GFP-RpL10A was driven in a small subpopulation of about 200 neurosecretory cells in the pars intercerebralis. TRAP-purified RNA was then compared to mRNA extracted from whole heads. They found that translated mRNAs encoding a neuropeptide specific to a small cell population of neurosecretory cells in the adult brain were enriched while another neuropeptide that is not expressed in these cells was depleted. Further, dllp2 transcript, which is expressed in only seven neurons within the pars intercerebralis, showed 55-fold enrichment in the affinity purified sample from pars intercerebralis neurons. In contrast, another gene, NPF, which is not expressed in the pars intercerebralis, was about 80-fold depleted. It has not been shown what percentage of expressing cells compared to total cells are required for obtaining optimal results to ensure that enrichment of transcripts can be accurately measured.
Recently, TRAP was performed in zebrafish, expressing the fusion protein EGFP-RpL10a under the control of the melanocyte-specific tyrosine related protein 1 promoter [31]. These experiments showed that melanocyte-specific genes are enriched in a zebrafish melanocyte-specific cell line as suggested by qPCR results. The differences of normalized Ct values (threshold cycle, the fractional cycle number at which the fluorescence passes the threshold) before and after TRAP purification were 2.5-5, showing that TRAP allowed isolation of cell-type-specific genes. Therefore, as suggested in this pilot study, TRAP is suitable to isolate genes from a small number of melanocyte-specific cells in zebrafish. More experiments are necessary to establish TRAP in zebrafish by showing differential gene expression profiles in a larger number of genes, ideally by performing RNA-seq.
Another study in zebrafish used TRAP for identification of factors that are required for heart regeneration after induced injury [70]. Among other genes that were differentially expressed (138 genes), the authors found that several members of the Janus kinase 1/Signal transducer and activator of transcription 3 (Jak1/Stat3) pathway showed elevated levels one day after injury compared to uninjured cardiomyocytes on microarray heat maps. They confirmed the up-regulation of four members of the Jak1/Stat3 pathway by qPCR, showing a 5 to 9-fold higher relative expression after injury compared to uninjured samples [70], suggesting that the genes that were identified as being differentially expressed in the TRAP microarray approach were indeed differentially expressed. Therefore, the TRAP method might be of great value in analyzing disease model systems.
Possible Explanations for High Background Levels
Similar to other RNA profiling methods, TRAP can have background contamination problems [29, 30]. Doyle et al. mentioned that a small number of probe sets were consistently enriched in every dataset analyzed, even in non-transgenic control mice, which they eliminated from all their datasets [29]. The authors suggested that some mRNAs might interact with monoclonal antibodies or protein G dynabeads in the absence of the EGFP fusion protein. Further, it appears that some transgenic mouse lines resulted in more background contamination than others [20, 71, 72], likely depending on which cell types were targeted and in how many cells the transgene was expressed, as suggested by the authors. Another contributor to non-specific transcripts is that small populations of unlabeled polysomes may be pulled down during the affinity purification step or that some mRNA transcripts are so abundant that they stick to the column and contaminate the purified sample.
concluding Remarks to TRAP
In summary, TRAP isolates actively translated mRNAs and therefore reveals which proteins are potentially relevant at a certain time in specific cell types. Even small cell populations expressing the fusion protein are suitable for translatome analyses. Several recent studies have now demonstrated that TRAP works well in various animal models. Interestingly, both TRAP and 4tU-labeling gene profiling studies in mouse have been performed using the Tie2 promoter [50, 68]. These two studies cannot be directly compared to each other because one method describes newly synthesized and the other method newly transcribed RNA. However, the data obtained from those very different methods that target distinct RNA populations, show some overlap of genes enriched in heart endothelial cells. This result suggests that, at least for some genes, increased translation results from increased transcription, and also confirms the specificity of the techniques used. It will be interesting to find similarities and differences between newly transcribed and currently translated mRNAs, identified by TRAP and 4tU-labeling, respectively, to better understand gene regulatory processes within specific cells. A comparison of data sets could identify whether enrichment in some transcripts correlates with a higher rate of translation or, whether differences in the pool of currently translated mRNAs can be traced back to increased transcription of a certain gene at a given time point. This comparison could be especially relevant in experiments analyzing the immediate effects of drug treatment.
NUCLEAR RNA PROFILING (INTACT)
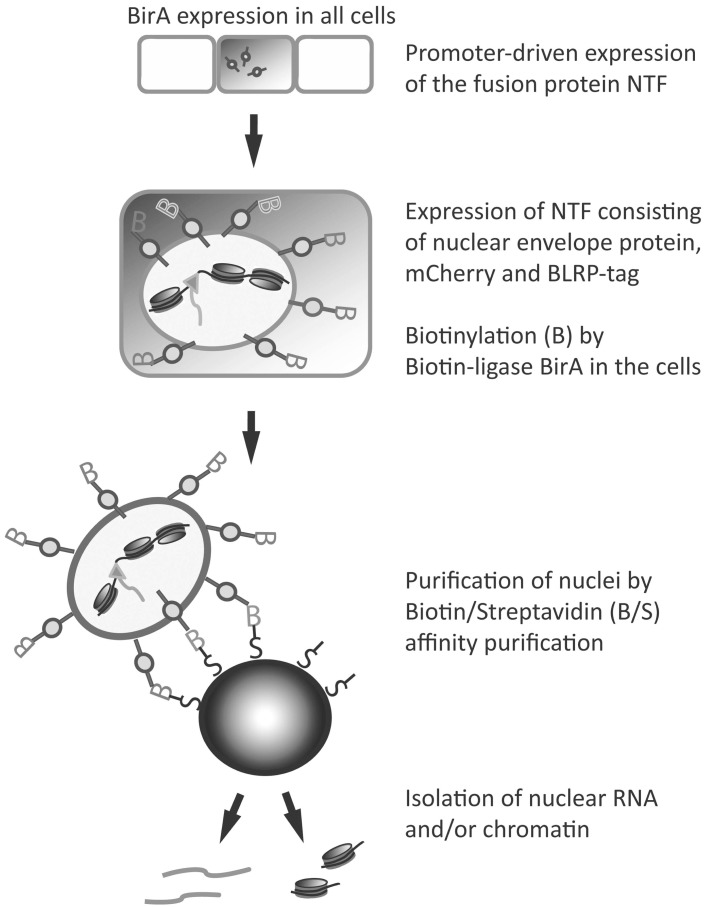
A sophisticated approach to better understand transcriptional regulation is to distinguish between nuclear and cytoplasmic RNA. The localization of transcripts within the cell, namely nuclear or cytoplasmic, might help us understand not just the expression patterns but also the regulation of these specific mRNAs. INTACT (isolation of nuclei tagged in specific cell types, Fig. 4), allows both nuclear RNA (nucRNA) profiling, which identifies all RNA within the nucleus and chromatin profiling, which identifies target loci of chromatin proteins [27]. INTACT is based on the analysis of nuclear RNA-seq and is often used in combination with chromatin immunoprecipitation (ChIP) [73], followed by sequencing (ChIP-seq).
Fig. (4).
Schematic overview of the method INTACT.
INTACT (Fig. 4) allows nucRNA and chromatin analysis in a cell-specific manner by expressing a nuclear targeting fusion (NTF) protein selectively in the nuclear envelope of specific cell types [27, 33]. In a study first performed in Arabidopsis, this NTF construct consisted of a fusion of three protein domains: an Arabidopsis-specific nuclear envelope targeting sequence (WPP domain of RAN GTPase activating protein 1, RanGAP1) for nuclear envelope targeting, green fluorescent protein (GFP) for visualization, and the biotin ligase recognition peptide (BLRP), a substrate for Escherichia coli biotin ligase (BirA), for nuclear pulldown [27]. Thus, this is a modular approach, with BirA biotinylated nuclei found only in cells expressing transgenes for two proteins, the NTF carrying the biotin ligase recognition peptide and in addition the BirA biotin ligase that biotinylates the NTF protein. Deal and Henikoff [27, 33] showed that biotinylated nuclei were isolated from Arabidopsis hair cells and non-hair cells of the root epidermis using biotin-streptavidin purification, leading to high yield and purity of nuclei specific to the cell type in which the fusion protein was expressed.
Using INTACT therefore allows isolation of cell-type specific nuclear RNA without the need to apply any substrates to induce transgene expression. Furthermore, the quality of the purification can be visually monitored by observing the ratio of fluorescently labeled to unlabeled nuclei before continuing with time-consuming and costly purification and sequencing steps.
Limitations of INTACT
The correlation of gene expression patterns of nuclear RNA and cellular RNA is controversial. In general, the abundance patterns of nuclear RNA, cytoplasmic, and total cellular mRNA are largely comparable, as demonstrated by a comparison of transcripts of the nuclear and the cytoplasmic compartments. In a study where the contributions of the nuclear and cytoplasmic compartments to global gene expression were compared in human cells, only 3% of all transcripts were found at significantly different levels, suggesting that nuclear and cytosolic total cellular RNA fractions can be equally used for gene expression level measurements [74]. The authors state that this generalization, however, is not applicable for transcripts encoding proteins associated with specific nuclear functions. Transcripts encoding proteins involved in cell cycle, mitosis, and transcription were overrepresented in nuclear RNA samples with respect to whole genome while transcripts for proteins regulating cell cycle, DNA replication, and DNA repair were overrepresented in the cytosol, compared to whole genome. Further, it is unclear whether nuclear RNA and cytoplasmic RNA is equivalent in all cell types during all stages of development and more studies are required to investigate differences in nuclear and cytoplasmic RNA levels.
In another report, Steiner et al. demonstrated strong correlation (R = 0.96) in the expression profiles between total cellular and nuclear RNA pools, confirming previously published results [75]. One caveat might be that, because nuclear RNA populations contain introns, RNA-seq read alignments hitting exons occurred less frequently in nuclear RNA samples [76] (on average about 75%) compared to whole-cell RNA samples (91%), suggesting that more RNA-seq reads would be necessary to achieve a similar exon coverage between nuclear and cytosol samples. In addition, the availability of sufficient starting material is a concern due to a yield of only 50-70% of the theoretically expected amounts of labeled nuclei [33]. Further, optimization of methods to isolate high quality intact nuclei from certain tissues might be required.
Efficiency of INTACT
As mentioned above, INTACT depends on high quality sample preparation and high purity of the sample because contamination with untagged nuclei would lead to high background noise. Sample quality, as determined by high purity (>95%) and high yield, allowed gene expression analysis in Arabidopsis root epidermis, resulting in the detection of 19 out of 24 known hair cell-specific genes in the hair cell sample and none in the non-hair sample, demonstrating that about 80% of the known hair cell markers were detected. In this study, a gene was defined as preferentially expressed in a given cell type if it showed an at least 1.3-fold difference between cell types with a Bayes p value of <0.02. Overall, 946 genes were found to be enriched in hair cells and 118 genes enriched in non-hair cells [27].
INTACT in Other Animal Models: C. elegans and Drosophila
Recently, Steiner et al. [75] successfully introduced use of the INTACT method for Caenorhabditis elegans (C. elegans) and Drosophila. Because the NTF protein described in Arabidopsis [27, 33] allows nuclear tagging only in Arabidopsis, new NTF proteins that are spatially accessible to biotinylation were designed specifically for each additional model system because each species requires a species-specific NTF protein that is designed to target the protein to the outer nuclear envelope. The C. elegans NTF is a chimera consisting of the C. elegans nuclear pore complex protein NPP-9 (a member of the nucleoporin family for localization to the cytoplasmic filaments of the nuclear pore), mCherry for visualization, a 3xFLAG tag for immunodetection, and a biotin ligase recognition peptide (BLRP), a preferred target for the biotin ligase BirA, necessary for biotinylation.
To target nuclei in Drosophila melanogaster, a fly-specific NTF was generated with the GTPase-activating protein RANGAP protein for targeting to the nuclear envelope [27], mCherry for visualization, the biotin ligase recognition peptide BLRP and a 3xFLAG tag for immunolabeling. This fly NTF was expressed under the control of the twist promoter to target and purify mesodermal nuclei.
Efficiency of INTACT in C. elegans and Drosophila
In C. elegans, nuclei expressing NTF-BirA in muscles, driven by the myo-3 promotor, were compared to nuclei expressing NTF only. The data sets, which showed a purity of >90% of nuclei expressing the fusion protein, were analyzed using whole-genome tilling microarrays, and showed that genes expressed in muscles were enriched while genes found in intestines and germ lines showed a tendency to be depleted. Averages of regularized t-values (based on a Bayesian probabilistic framework for microarray data analysis, using the program Cyber-T [77]) are around five for muscle-specific genes, whereas t-values for intestine- or germline-specific genes are about -1 or -2, suggesting a more than 5-fold enrichment of muscle-specific genes compared to intestine- or germline-specific genes. Further, 71% of the 200 most enriched genes in the microarray data set were found to be expressed in muscle cells while 77% of the 200 most depleted genes were found to be expressed in non-muscle cells [75].
To verify INTACT results in Drosophila, the relative transcript level of four mesoderm-specific genes was measured by qPCR in total nuclei and affinity-purified nuclei. Three of the four selected mesodermal genes showed enrichment [75] with relative expression levels of 9 to 17-fold for affinity-purified RNA relative to total RNA. This case study suggests that INTACT will allow the identification of gene expression profiles in specific cells in the fly.
Modified INTACT Method Using a Different Protein Tag
As an alternative method to tag nuclei for isolation in Drosophila, Henry et al. [76] modified the INTACT method by using an NTF fusion protein based on the C. elegans protein UNC-84. This member of the SUN domain family (named for the C. elegans proteins Sad1p and UNC-84 domains) is embedded in the inner nuclear membrane of all eukaryotes and is thought to be involved in the positioning of the nucleus in the cell. The localization of the tag in the inner nuclear envelope is of advantage because the authors use a more efficient nuclei isolation technique that breaks up the outer nuclear envelope but leaves the inner nuclear envelope intact. The authors made two different constructs, one fusing two copies of GFP to UNC-84 to make a green tag and one fusing tdTomato to a 3xFLAG tag to make a red tag. These tags allow immunoaffinity purification compared to the biotin-streptavidin purification and appear to result in comparable purity and yield. Using GFP or FLAG tags allow the researcher more flexibility since both tags could be expressed simultaneously in different tissues within one organism and then purified independently using antibodies to either GFP or FLAG.
One advantage of the GFP- or FLAG-tag-based system is that only the NTF construct needs to be expressed in the cell-type of interest, while with the INTACT method described earlier, both the NTF protein and a BirA protein need to be expressed simultaneously in order to enable cell-specific biotin-streptavidin based purification. Nuclei containing either GFP- or FLAG- tag were isolated using immunoaffinity purification with either anti-GFP or anti-FLAG coated magnetic beads, respectively. The use of the Gal4/UAS system allowed the authors to express their nuclear tags in different tissues, depending on available driver lines.
Efficiency of the Modified INTACT Method
The modified method described by Henry et al. [76] allowed the analysis of three different Drosophila neuronal cell populations that expressed the UAS-nuclear tag cassettes broadly in roughly 100 000 brain cells (pan-neuronal driver), in about 2000 brain cells (Kenyon cells), or in about 130 cells per brain (octopaminergic neurons). Differential expression profiles were described in the context of known marker genes. In general, the gene expression that was found in the different populations correlated well with known gene expression patterns. A more detailed comparison of gene expression levels of the two neuronal subpopulations, the Kenyon cells and the octopaminergic neurons, showed that nuclei from the Kenyon cell population were 5.5-fold enriched for a vesicular transporter gene known to be expressed in the mushroom body. Nuclei from octopaminergic neurons showed a 58-fold and 30-fold enrichment of the biosynthetic enzymes Tdc2 and Tbh (genes encoding enzymes for octopamine synthesis), respectively, compared to pan-neuronal nuclei. Other genes involved in the glutamate pathway that were previously described to be expressed in octopaminergic neurons were moderately enriched with 2.7 and 1.7-fold higher expression levels. Thus, the efficiency of the modified INTACT method is comparable to the original INTACT method, but with greater versatility afforded by the simplified immunoprecipitation and Gal4/UAS expression system.
Background Problems
Similar to other purification methods based on immunoprecipitation, some background contamination is found. Deal et al. [33] describe a purity of 90-98% under optimal conditions, meaning that at least 90% of the nuclei carry the transgene. Purity of isolated nuclei is critical for the outcome of the experiment and is built on a high-quality isolation of intact nuclei, because the integrity of nuclear envelopes, either inner or both layers depending on the NTF protein used, is important for the purification of NTF-labeled nuclei. One possible source of contamination is cell debris and unlabeled nuclei that are not sufficiently removed from the lysate. Further, when purifying rare cell types, more contamination might be expected because more tissue must be purified to obtain a sufficient amount of labeled nuclei.
concluding Remarks to INTACT
In summary, INTACT uses deep sequencing of nuclear RNAs (nucRNA-seq), representing unspliced and spliced transcripts in the nucleus and results are thought to correlate with primary cytosolic transcript frequencies for most transcripts. Some exceptions apply for genes important for general cellular processes, which appear to be overrepresented in nuclear RNA. In general, both INTACT and 4tU-labeling methods could be used to identify RNA populations within specific cells. One advantage of INTACT over 4tU-labeling method is that the purity of nuclei expressing the fusion protein can be visually monitored, although background problems due to insufficient sample purity were mentioned for some tissues. Further, INTACT, contrary to 4tU-labeling, does not require supplement with any substrate, which might be of advantage in tissues that are difficult to penetrate.
CHROMATIN PROFILING (CHIP)
Epigenomic profiling by chromatin immunoprecipitation sequencing (ChIP-seq) can be performed in parallel to nucRNA sequencing to identify actively transcribed genes, putatively regulatory regions, and ncRNAs, depending on the ChIP analysis of choice [73]. Epigenomic profiling analyzes the control of gene expression and is used in the context of nucleosomes, the basic units of chromatin, which bind specific DNA sequences and thereby regulate access of proteins that control transcription and replication to the DNA [78]. Histones form the core of nucleosomes and chromatin immunoprecipitation (ChIP) can detect protein-DNA binding interactions as well as modified histone proteins. By combining nuclear RNA-seq with chromatin profiling, information can be gained about gene expression and gene regulation by analyzing nucleosome occupancy.
Many different DNA-binding proteins are necessary to fulfill basic cellular processes such as transcription, splicing, replication and more. In addition, a combination of chromatin-based mechanisms involving transcription factor binding, nucleosome remodeling, deposition of histone variants, and post-translational histone modifications are involved in these processes [73, 79, 80]. Different experimental conditions allow either detection of protein binding sites onto DNA (DNA-binding protein ChIP-seq), or identification of chemical histone modifications (Histone modification ChIP-seq). Furthermore, regions of nucleosome-depleted open chromatin can be identified using DNase-seq or formaldehyde-assisted identification of regulatory elements (FAIRE) [73]. These methods can predict where potential binding sites are located but cannot identify which specific factors are bound to the DNA.
ChIP Analysis in Arabidopsis
The addition of chromatin analyses for two different histone methylation modifications allowed Deal and Henikoff [27] to confirm the accuracy of gene expression profiles obtained by INTACT [27]. For histone modification ChIP, the authors used the transcription-associated mark trimethylation of lysine 4 in histone-3 (H3K4me3), which is associated with highly expressed genes, and the Polycomb silencing-associated mark trimethylation of lysine 27 in histone-3 (H3K27me3), which is excluded from the most highly expressed genes. In an attempt to correlate gene expression with each modification, the authors confirmed that most genes with differential gene expression could be correlated with changes in the balance of these two chromatin modifications in a given cell type. In genes with attenuated expression levels, the level of H3K4me3 within a gene is decreased and the level of H3K27me3 modification is increased.
ChIP Analysis in C. elegans and Drosophila
Steiner et al. [75] used their C. elegans sample for performing nucleosome occupancy profiles of total and affinity-purified nuclei using micrococcal nuclease, an endo-exonuclease that makes double-stranded cuts between nucleosome particles, followed by paired-end sequencing of genomic DNA. Genes that showed the highest expression in muscle nuclei compared with total nuclei could be correlated with a reduction in nucleosome occupancy over their promoters and coding sequences.
In their proof of principle study, Henry et al. described both the profiling of gene expression followed by RNA-seq and the distribution of differential histone modifications that indicate active promotors (H3K4me3), open chromatin (H3K27ac), and polycomb group (PcG)-mediated transcriptional silencing (H3K27me3) [76]. The authors observed strongly opposing differential H3K27me3 and H3K27ac signals in octopaminergic neurons versus Kenyon cells and established a correlation between differential histone modifications and differentially expressed genes in both neuronal populations, suggesting that INTACT has the ability to identify transcriptional regulatory networks that underlie cell identity.
Recent advances in ChIP methodology make this challenging assay more accessible and allow ChIP analyses for broader sets of experiments. For instance, now only 10,000 cells, instead of millions of cells, are required for histone modification experiments, allowing more different specific cell types to be analyzed for protein-DNA interactions (see Furey [73]). ChIP is the only method that can identify DNA-binding proteins or modified histones genome-wide, making it an important resource for understanding transcriptional regulation and other biological processes in a specific cell type.
CONCLUSION
The different RNA isolation methods described in this review allow selective isolation of biologically relevant mRNA populations at an investigator-selected time point (Fig. 1, Table 3). Thiol-labeling might be the method of choice when genes need to be identified that are up-regulated or down-regulated in certain tissues or that are differentially expressed in disease models compared to wild types. Alternatively, if one is interested in nuclear rather than cytoplasmic or whole-cell RNAs, then the INTACT method can help identify differentially expressed genes at a certain time point. If, on the other hand, one’s interest is to identify genes that are actively translated at a given time, then polyribosome isolation methods should be chosen to highlight the formation of newly-synthesized proteins in specific tissues. In general, most techniques described in this review are fairly new and many of the reports are proof-of-principle studies. As these methods are put into further use, it will become more possible to evaluate the balance of benefits and drawbacks of each technique.
The techniques described above promise to have a major impact in the understanding of gene-regulatory networks by unraveling changes in developmental gene expression profiles in different cell types over time. In addition, comparing data sets using a method that identifies newly synthesized versus currently translated mRNAs will allow us to better understand the complicated regulatory machinery that controls gene transcription and selects mRNAs for translation. Future studies combining transcriptional and translational profiling with proteomics will complete the big picture of gene regulation and expression. Building an open access gene expression database would be extremely useful and would allow direct comparisons of different data sets. Such a database would have an enormous potential for unraveling the processes involved in establishing cell identity during development and cell function for physiology. Furthermore, transcriptional and translational profiling could aid in the discovery and development of biomarkers for human diseases [81]. A significant area that must be explored in more detail is the differential expression of all types of ncRNA because they appear to be important players in the regulation of gene expression.
ACKNOWLEDGEMENTS
This work was supported by R24 OD011199 and R01 OD011116 (R01 RR020833) from the Office of the Director, National Institutes of Health (J.H.P.), by R01NS065795 from the National Institute of Neurological Disorders and Stroke (P.W.), and by 5 P01 HD22486 from the Eunice Kennedy Shriver National Institute of Child Health and Development (P.W. and J.H.P.).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Esteller M. Non-Coding Rnas In Human Disease. Nat. Rev. Genet. 2011;12(12): 861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 2.Tuck A C, Tollervey D. Rna In Pieces. Trends Genet. 2011;27 (10):422–432. doi: 10.1016/j.tig.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Mercer T R, Mattick J S. Structure And Function Of Long Noncoding Rnas In Epigenetic Regulation. Nat. Struct. Mol. iol. 2013;20 (3):300–7. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 4.Mercer T R, Dinger M E, Mattick J S. Long Non-Coding Rnas Insights Into Functions. Nat. Rev. Genet. 2009;10 (3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Dinger ME., Pang K. C., Mercer T. R., Mattick J. S. Differentiating Protein-Coding And Noncoding Rna Challenges And Ambiguities. Plos Comput. Biol. 2008;4 (11):E1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lodish H B. A, Zipursky S L, editors. 4th Ed. New York.: Free-Man ; 2000. ProCessing Of Rrna And Trna. [Google Scholar]
- 7.Kim V N, Han J, Siomi M C. Biogenesis Of Small Rnas In Animals. Nat. Rev. Mol. Cell Biol. 2009;10 (2): 126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V, Chen X. The Regulation Of Genes And Genomes By Small Rnas. Development. 2007;134 (9): 1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 9.Ishizu H, Siomi H, Siomi M C. Biology Of Piwi-Interacting Rnas New Insights Into Biogenesis And Function Inside And Outside Of Germlines. Genes Dev. 2012;26 (21):2361–2373. doi: 10.1101/gad.203786.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bratkovic T, Rogelj B. Biology And Applications Of Small Nucleolar Rnas. Cell. Mol. Life Sci. 2011;68 (23): 3843–51. doi: 10.1007/s00018-011-0762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalil A M, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein B E, Van Oudenaarden A, Regev A, Lander E S, Rinn J L. Many Human Large In-Tergenic Noncoding Rnas Associate With Chromatin-Modifying Complexes And Affect Gene Expression. Proc. Natl. Acad. Sci. Usa. 2009;106 (28): 11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rinn J L, Chang H Y. Genome Regulation By Long Noncoding Rnas. Annu. Rev. Biochem. 2012;81 (1): 145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang E, Van Nimwegen E, Zavolan M, Rajewsky N, Schroeder M, Magnasco M, Darnell J E., Jr Decay Rates Of Hu-Man Mrnas Correlation With Func-Tional Characteristics And Sequence Attributes. Genome Res. 2003;13 (8):1863–72. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharova L V, Sharov A A, Nedorezov T, Piao Y, Shaik N, Ko. M S. Database For Mrna Half-Life Of 19 977 Genes Obtained By Dna Microarray Analysis Of Pluripotent And Differentiating Mouse Embryonic Stem Cells. Dna Res. 2009;16 (1): 45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shyu A B, Greenberg M E, Belasco J G. The C-Fos Transcript Is Targeted For Rapid Decay By Two Distinct Mrna Deg-Radation Pathways. Genes Dev. 1989;3 (1):60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 16.Dolken L, Ruzsics Z, Rädle B, Friedel C C, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, Koszinowski U H. High-Resolution Gene Expression Profiling For Simultaneous Kinetic Parameter Analysis Of Rna Synthesis And Decay. Rna. 2008;14 (9): 1959–1972. doi: 10.1261/rna.1136108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Windhager L, Bonfert T, Burger K, Ruzsics Z, Krebs S, Kaufmann S, Mal-Terer G, L'hernault A, Schilhabel M, Schreiber S, Rosenstiel P, Zimmer R, Eick D, Friedel C C, Dolken L. Ultra-Short And Progressive 4su-Tagging Re-Veals Key Characteristics Of Rna Pro-Cessing At Nucleotide Resolution. Genome Res. 2012;22 (10): 2031–42. doi: 10.1101/gr.131847.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. Rna-Seq A Revolutionary Tool For Tran-Scriptomics. Nat. Rev. Genet. 2009;10 (1): 57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki E S. Microarrays And The Gene Expression Profile Of A Single Cell. Ann. N. Y. Acad. Sci. 2004;1020:92–100. doi: 10.1196/annals.1310.010. [DOI] [PubMed] [Google Scholar]
- 20.Okaty B W, Sugino K, Nelson S B. Cell Type-Specific Transcriptomics In The Brain. J. Neurosci. 2011;31 (19):6939–43. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Li. Y, Fan X, Zhang Y, Wu. J, Zhao Z. Early Proliferation Alteration And Differential Gene Expression In Human Periodontal Ligament Cells Subjected To Cyclic Tensile Stress. Arch. Oral Biol. 2011;56 (2):177–186. doi: 10.1016/j.archoralbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Cleary M D, Meiering C D, Jan E, Guymon R, Boothroyd J C. Biosyn-Thetic Labeling Of Rna With Uracil Phosphoribosyltransferase Allows Cell-Specific Microarray Analysis Of Mrna Synthesis And Decay. Nat. Bio-Technol. 2005;23 (2):232–7. doi: 10.1038/nbt1061. [DOI] [PubMed] [Google Scholar]
- 23.Kenzelmann M, Maertens S, Hergen-Hahn M, Kueffer S, Hotz-Wagenblatt A , Li L, Wang S, Ittrich C, Lemberger T, Arribas R, Jonnakuty S, Hollstein M C, Schmid W, Gretz N J G H G S. Mi-Croarray Analysis Of Newly Synthe-Sized Rna In Cells And Animals. Proc. Natl. Acad. Sci. Usa. 2007;104 (15):6164–6169. doi: 10.1073/pnas.0610439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller M R, Robinson K J, Cleary M D, Doe C Q. Tu-Tagging Cell Type-Specific Rna Isolation From Intact Complex Tissues. Nat. Methods. 2009;6 (6):439–41. doi: 10.1038/nmeth.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeiner GM., Cleary M.D, Fouts AE., Meiring C.D, Mocarski E S, Boothroyd J C. Rna Analysis By Biosynthetic Tagging Using 4-Thiouracil And Uracil Phosphoribosyltransferase. Methods Mol. iol. 2008;419:135–46. doi: 10.1007/978-1-59745-033-1_9. [DOI] [PubMed] [Google Scholar]
- 26.Sanz E, Yang L, Su. T, Morris DR., Mckight G. S., Amieux P. S. Cell-Type-Specific Isolation Of Ribosome-Associated Mrna >From Complex Tissues. . Proc. Natl. Acad Sci. USA . 2009;106 (33):13939–13944. doi: 10.1073/pnas.0907143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deal R B, Henikoff S . Simple Method For Gene Expression And Chromatin Profiling Of Individual Cell Types Within A Tissue. Dev. Cell. 2010;18 (6):1030–40. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleary M D. Chapter 19 Cell-Specific Analysis Of Mrna Synthesis And Decay In Vivo With Uracil Phosphoribosyl-Transferase And 4-Thiouracil. In: Methods Enzymol., Lynne E. M., Megerditch K., editors. Vol. Volume 448. Academic Press.; 2008. pp. 379–406. [DOI] [PubMed] [Google Scholar]
- 29.Doyle J P, Dougherty J D, Heiman M, Schmidt E F, Stevens T R, Ma. G, Bupp S, Shrestha P, Shah R D, Doughty M L, Gong S, Greengard P, Heintz N. Ap-Plication Of A Translational Profiling Approach For The Comparative Analysis Of Cns Cell Types. Cell. 2008;135 (4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiman M, Schaefer A, Gong S, Peter-Son J D, Day M, Ramsey K E, Suárez-Fariñas M, Schwarz C, Stephan D A, Surmeier D J, Greengard P, Heintz N. A Translational Profiling Approach For The Molecular Characterization Of Cns Cell Types. Cell. 2008;135 (4): 738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tryon R C, Pisat N, Johnson S L, Dougherty J D. Development Of Translating Ribosome Affinity Purifi-Cation For Zebrafish. Genesis. 2013;51 (3):187–192. doi: 10.1002/dvg.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanz E, Evanoff R, Quintana A, Evans E, Miller J A, Ko C, Amieux P S, Griswold M D, Mcknight G S. Ribotag Analysis Of Actively Translated Mrnas In Sertoli And Leydig Cells In Vivo. Plos One . 2013;8 (6):E66179. doi: 10.1371/journal.pone.0066179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deal R B, Henikoff S. The Intact Method For Cell Type-Specific Gene Ex-Pression And Chromatin Profiling In Arabidopsis Thaliana. Nat. Prtoc. 2011; 6 (1):56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchell J A, Clay I, Umlauf D, Chen C Y, Moir C A, Eskiw C H, Schoenfelder S, Chakalova L, Nagano T, Fraser P. Nuclear Rna Sequencing Of The Mouse Erythroid Cell Transcriptome. Plo One. 2012;7 (11):E49274. doi: 10.1371/journal.pone.0049274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simone N L, Bonner R F, Gillespie J W, Emmert-Buck MR., Lotta L. A. Laser-Capture Microdissec-Tion Opening The Microscopic Frontier To Molecular Analysis. . Trends Genet. . 1998; 14 (7):272–276. doi: 10.1016/s0168-9525(98)01489-9. [DOI] [PubMed] [Google Scholar]
- 36.Mach W, Thimmesch A, Orr J, Slusser J, Pierce J. Flow Cytometry And Laser Scanning Cytometry, A Comparison Of Techniques. J. Clin. Monit. Coput. 2010;24 (4):251–259. doi: 10.1007/s10877-010-9242-4. [DOI] [PubMed] [Google Scholar]
- 37.Lipovsek D, Plückthun A. In-Vitro Protein Evolution By Ribosome Display And Mrna Display. J. Immunol. Mehods. 2004;290 (1 2):51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 38.Cha S. Development Of Inhibitors Of Pyrimidine Metabolism. Yonsei Me. J. 1989;30 (4):315–326. doi: 10.3349/ymj.1989.30.4.315. [DOI] [PubMed] [Google Scholar]
- 39.Melvin W T, Milne H B, Slater A A, Allen H J, Keir H M. Incorporation Of 6-Thioguanosine And 4-Thiouridine Into Rna. Eur. J. Biochem. 1978;92 (2): 373–379. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- 40.Johnson T R, Rudin S D, Blossey B K, Ilan J, Ilan J. Newly Synthesized Rna Simultaneous Measurement In Intact Cells Of Transcription Rates And Rna Stability Of Insulin-Like Growth Fac-Tor, I.ctin And Albumin In Growth Hormone-Stimulated Hepatocytes. Proc. Natl. Acad. Sci. U S A. . 1991; 88 (12):5287–91. doi: 10.1073/pnas.88.12.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donald R. G. K, Roos D, Ullman B. Molecular And Biochemical Parasit-Ologycarter, D Expression, Purification, And Characterization Of Uracil Phos-Phoribosyltransferase From Toxo-Plasma Gondii. Mol. Biochem. Parasitol. 1997;87 (2): 137–144. doi: 10.1016/s0166-6851(97)00058-3. [DOI] [PubMed] [Google Scholar]
- 42.Iltzsch M H. Pyrimidine Salvage Path-Ways In Toxoplasma Gondii. J. Eukaryot. Microbiol. 1993;40 (1): 24–28. doi: 10.1111/j.1550-7408.1993.tb04877.x. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Huang S, Chen J, Yang Z, Fei X, Zheng M, Ji. C, Xie Y, Mao Y. Identifi-Cation And Characterization Of Human Uracil Phosphoribosyltransferase (Uprtase) . J. Hum. Genet. 2007;52 (5): 415–22. doi: 10.1007/s10038-007-0129-2. [DOI] [PubMed] [Google Scholar]
- 44.Brand A H, Perrimon N. Targeted Gene Expression As A Means Of Altering Cell Fates And Generating Dominant Phenotypes. Development. 1993;118 (2): 401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 45.Southall T D, Elliott D A, Brand A H. The Gal4 System A Versatile Toolkit For Gene Expression In Drosophila. Cold Spring Harbor Protocols . 2008;(7):Pdb.Top49. doi: 10.1101/pdb.top49. [DOI] [PubMed] [Google Scholar]
- 46.Davison J M, Akitake C M, Goll M G, Rhee J M, Gosse N, Baier H, Halpern M E, Leach S D, Parsons M J. Trans-Activation >From Gal4-Vp16 Transgenic Insertions For Tissue-Specific Cell La-Beling And Ablation In Zebrafish. Dev. Biol. 2007;304 (2): 811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halpern M E, Rhee J, Goll M G, Akitake C M, Parsons M, Leach S D. Gal4/Uas Transgenic Tools And Their Application To Zebrafish. Zebrafish. 2008;5 (2): 97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewandoski M. Conditional Control Of Gene Expression In The Mouse. Nat. Rev. Genet. 2001;2 (10):743–55. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 49.Orban P C, Chui D, Marth J D. Tissue- And Site-Specific Dna Recombination In Transgenic Mice. Proc. Natl. Acad. Sci. U S A. 1992;89 (15): 6861–5. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gay L, Miller M R, Ventura P B, Dev-Asthali V, Vue Z, Thompson H L, Tem-Ple S, Zong H, Cleary M D, Stankunas K, Doe C Q, Mouse u. Tagging A Chem-Ical/Genetic Intersectional Method For Purifying Cell Type-Specific Nas-Cent Rna. Genes Dev. 2013;27 (1): 98–115. doi: 10.1101/gad.205278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleary M D. Cell Type-Specific Analysis Of Mrna Synthesis And Decay In Vivo With Uracil Phosphoribosyltrans-Ferase And 4-Thiouracil. Methods En-Zymol. 2008;448:379–406. doi: 10.1016/S0076-6879(08)02619-0. [DOI] [PubMed] [Google Scholar]
- 52.Friedel C C, Dolken L. Metabolic Tagging And Purification Of Nascent Rna Implications For Transcriptomics. Mol. Biosystems. 2009;5 (11): 1271–8. doi: 10.1039/b911233b. [DOI] [PubMed] [Google Scholar]
- 53.Rabani M, Levin J Z, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Metabolic Labeling Of Rna Uncovers Principles Of Rna Production And Degradation Dynamics In Mammalian Cells. Nat. Biotechnol. 2011;29 (5): 436–42. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J, Sun M, Kent W J, Huang X, Xie H, Wang W, Zhou G, Shi RZ., Rowley J. D. Over 20% Of Human Transcripts Might Form Sense Antisense Pairs. Nucleic Acids Res. . 2004;32 (16): 4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reyes P, Guganig ME. Studies On A Py-Rimidine Phosphoribosyltransferase From Murine Leukemia P1534j. Partial Purifiction Substrate Specificity, And Evidence For Its Existence As A Bifunc-Tional Complex With Orotidine 5-Phosphate Decarboxylase. J. Biol. Chem. . 1975;250 (13): 5097–108. [PubMed] [Google Scholar]
- 56.Carter D, Donald R G, Roos D, Ullman B. Expression, Purification, And Characterization Of Uracil Phos-Phoribosyltransferase >From Toxo-Plasma Gondii. Mol. Biochem. Parasitol. 1997;87 (2): 137–44. doi: 10.1016/s0166-6851(97)00058-3. [DOI] [PubMed] [Google Scholar]
- 57.Payne R C, Cheng N, Traut T W. Uridine Kinase From Ehrlich Ascites Carcinoma.Purification And Properties Of Homogeneous Enzyme. J. Biol. Chem. 1985;260 (18): 10242–7. [PubMed] [Google Scholar]
- 58.King A E, Ackley M A, Cass C E, Young J D, Baldwin S A. Nucleoside Transporters From Scavengers To Novel Therapeutic Targets. Trends Pharmacol. Sci. 2006;27 (8): 416–25. doi: 10.1016/j.tips.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Cansev M. Uridine And Cytidine In The Brain Their Transport And Utilization. Brain Res. Rev. 2006;52 (2):389–97. doi: 10.1016/j.brainresrev.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Del Prete MJ., Vrnal R., Dolznig H. M , llner E. W., Garcia-Sanz J. A. Isola-Tion Of Polysome-Bound Mrna From Solid Tissues Amenable For Rt-Pcr And Profiling Experiments. Rna . 2007; 13 (3): 414–421. doi: 10.1261/rna.79407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian Q, Stepaniants S B, Mao M, Weng L, Feetham M C, Doyle M J, Yi E C, Dai H, Thorsson V, Eng J, Goodlett D, Berger J P, Gunter B, Linseley P S, Stoughton R B, Aebersold R, Collins S J, Hanlon W A, Hood L E. Integrated Genomic And Proteomic Analyses Of Gene Expression In Mammalian Cells. Mol. Cell. Proteom. 2004;3 (10): 960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Gygi S P, Rochon Y, Franza B R, Aebersold R. Correlation Between Protein And Mrna Abundance In Yeast. Mol. Cell.iol. . 1999;19 (3): 1720–30. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kristensen A R, Gsponer J, Foster L J. Protein Synthesis Rate Is The Predomi-Nant Regulator Of Protein Expression During Differentiation. Mol. Syst.iol. . 2013; 9:689. doi: 10.1038/msb.2013.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dana A, Tuller T. Determinants Of Translation Elongation Speed And Ri-Bosomal Profiling Biases In Mouse Em-Bryonic Stem Cells. Plos Comput.iol. . 2012; 8 (11): E1002755. doi: 10.1371/journal.pcbi.1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arava Y, Boas F E, Brown P O, Herschlag D. Dissecting Eukaryotic Translation And Its Control By Ribo-Some Density Mapping. Nucleic Acids Res. 2005; 33 (8): 2421–32. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halbeisen R E, Gerber A P. Stress-Dependent Coordination Of Transcrip-Tome And Translatome In Yeast. Plos Biol. 2009;7 (5): E1000105. doi: 10.1371/journal.pbio.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guttman M, Russell P, Ingolia Nicho-Las T, Weissman Jonathan S, Lander Eric S. Ribosome Profiling Provides Ev-Idence That Large Noncoding Rnas Do Not Encode Proteins. Cell. 2013;154 (1): 240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou P, Zhang Y, Ma. Q, Gu F, Day D S, He A, Zhou B, Li J, Stevens S M, Romo D, Pu W T. Interrogating Translational Efficiency And Lineage-Specific Transcriptomes Using Ribosome Affinity Purification. Proc. Natl. Acad. Sci. Usa. 2013;110 (38): 15395–15400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas A, Lee P J, Dalton J E, Nomie K J, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN., Dierick H. A. A. Versatile Method For Cell-Specific Profiling Of Translated Mrnas In Drosophila. Plos One. 2012; 7 (7): E40276. doi: 10.1371/journal.pone.0040276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang Y, Gupta V, Karra R, Holdway J E, Kikuchi K, Poss K D. Translational Profiling Of Cardiomyocytes Identifies An Early Jak1/Stat3 Injury Response Required For Zebrafish Heart Regeneration. Proc. Natl. Acad. Sci. Usa. 2013;110 (33): 13416–13421. doi: 10.1073/pnas.1309810110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dougherty J D, Schmidt E F, Nakajima M, Heintz N. Analytical Approaches To Rna Profiling Data For The Identifi-Cation Of Genes Enriched In Specific Cells. Nucleic Acids Res. 2010;38 (13): 4218–4230. doi: 10.1093/nar/gkq130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okaty B W, Sugino K, Nelson S B. A Quantitative Comparison Of Cell-Type-Specific Microarray Gene Expression Profiling Methods In The Mouse Brain. Plos One. 2011;6 (1): E16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Furey T S. Chip-Seq And Beyond New And Improved Methodologies To Detect And Characterize Protein-Dna Interactions. Nat. Rev. Genet. 2012;13 (12): 840–52. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barthelson R A, Lambert G M, Vanier C, Lynch R M, Galbraith D W. Compar-Ison Of The Contributions Of The Nu-Clear And Cytoplasmic Compartments To Global Gene Expression In Human. Cells. Bmc Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steiner F A, Talbert P B, Kasinathan S, Deal R B, Henikoff S. Cell-Type-Specific Nuclei Purification From Whole Animals For Genome-Wide Expression And Chromatin Profiling. Genome Res. 2012;22 (4): 766–77. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Henry G L, Davis F P, Picard S, Eddy S R. Cell Type-Specific Genomics Of Dro-Sophila Neurons. Nucleic Acids Res. 2012;40 (19):9691–9704. doi: 10.1093/nar/gks671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baldi P, Long A D. A Bayesian Frame-Work For The Analysis Of Microarray Expression Data Regularized T -Test And Statistical Inferences Of Gene Changes. Bioinformatics. 2001;17 (6): 509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 78.Deal R B, Henikoff S. Capturing The Dynamic Epigenome. Genome Biol. 2010;11 (10): 218. doi: 10.1186/gb-2010-11-10-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ng R K, Gurdon J B. Epigenetic Inher-Itance Of Cell Differentiation Status. Cell Cycle. 2008;7 (9): 1173–7. doi: 10.4161/cc.7.9.5791. [DOI] [PubMed] [Google Scholar]
- 80.Yuan G, Zhu B. Histone Variants And Epigenetic Inheritance. Biochim. Bio-Phys. Acta. 2012;1819 (3-4): 222–9. doi: 10.1016/j.bbagrm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Macisaac K D, Wang I M, Menetski J, Roberts C. Genomic And Systems Ap-Proaches To Translational Biomarker Discovery In Immunological Diseases. Drug Discov. Today. 2014;19 (2): 133–139. doi: 10.1016/j.drudis.2013.10.002. [DOI] [PubMed] [Google Scholar]