Figure 1.
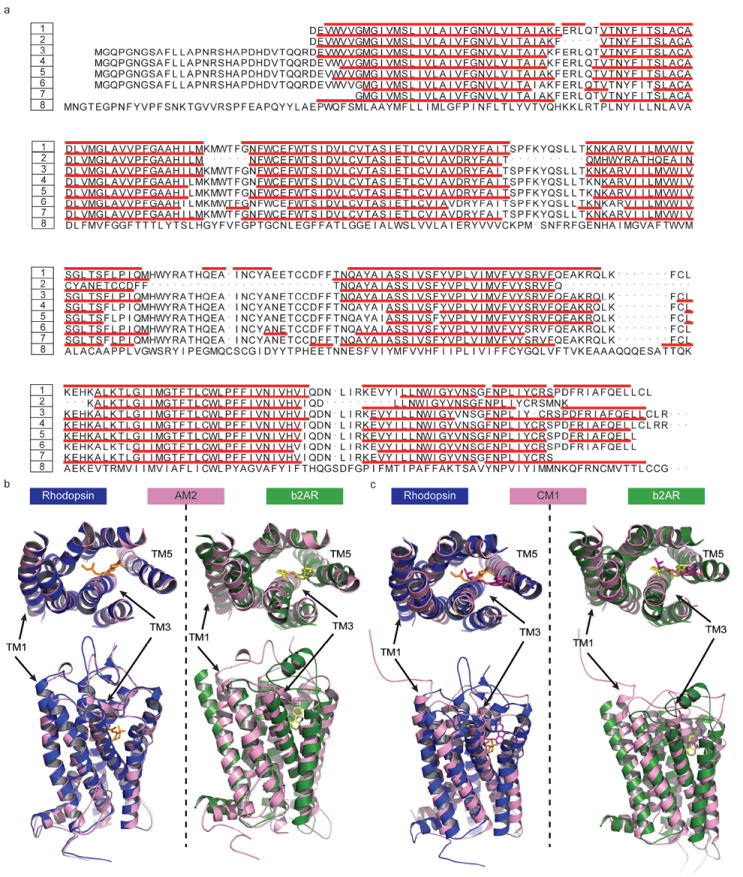
The structural similarity analysis of theoretical models in comparison with crystal structures. (a) The secondary structure assignment for TM segments of six theoretical models and two crystal structures (2RH1 and 1U19). The numbers and their corresponding structures are coded by 1:2RH1, 2:AM1, 3:AM2, 4:AM3, 5:CM1, 6:CM2, 7:CM3, 8:1U19. The remaining two models, i.e., AM4 and CM4, share similar backbone structures to CM3 with pairwise RMSD of TMs less than 0.4 Ǻ. Therefore only CM3 is included in the sequence alignment plot. The red bars indicate the helical structure elements identified by MOE. (b) The structural superposition of the theoretical models AM2 (rendered in pink) to 2RH1 (rendered in dark green) and 1U19 (rendered in blue). Note that the most structurally divergent TM regions are indicated. (c) The structural superposition of the theoretical models CM1 (rendered in pink) to 2RH1 (rendered in dark green) and 1U19 (rendered in blue). Note that the most structurally divergent TM regions are indicated as well.
