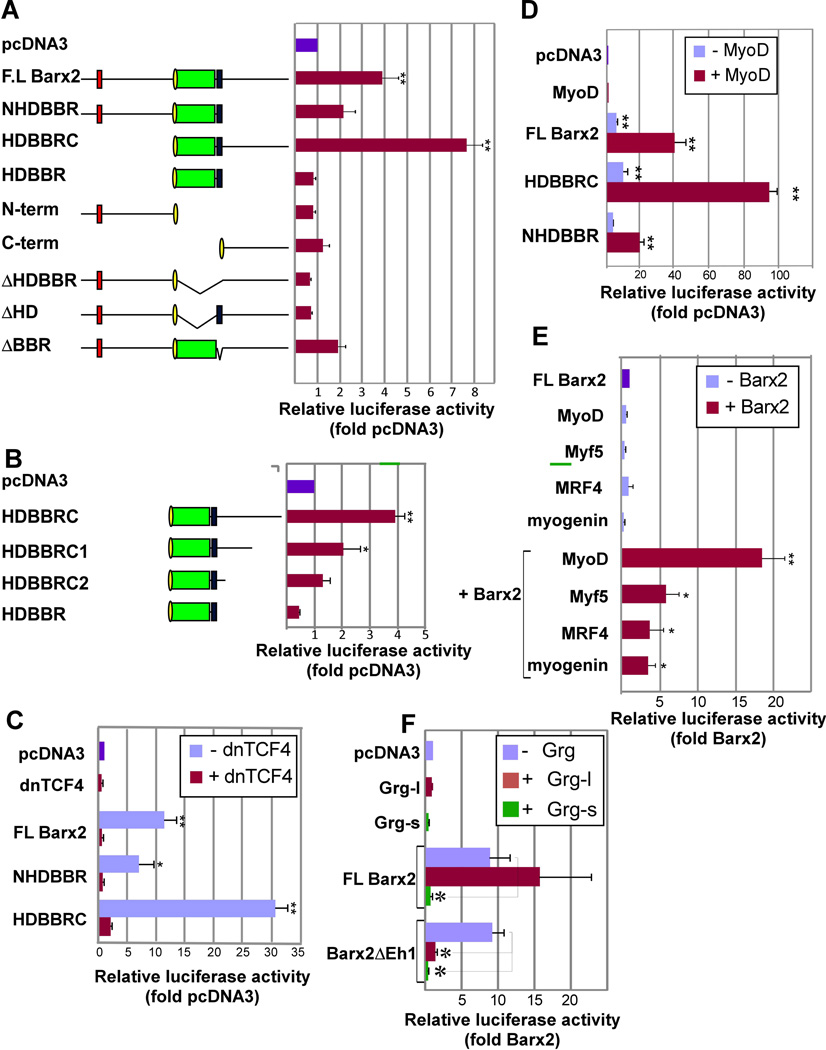
Figure 2.
Barx2 regulates the TOPflash reporter gene alone and synergistically with MRFs in C2C12 cells. A. Analysis of Barx2 deletion constructs shows that the homeodomain and BBR are required for activation of the TOPflash reporter gene. Left: schematic of Barx2 deletion constructs. Green – homeodomain; blue – BBR; yellow – nuclear localization sequence; red- Eh-1 motif. Right: TOPflash luciferase activity after cotransfection of Barx2 constructs in C2C12 cells. B. Analysis of Barx2 deletion constructs shows that the Barx2 activation function is distributed throughout the C-terminal domain. Left: schematic of Barx2 deletion constructs. Right: TOPflash luciferase activity. C. Co-transfection of Barx2 constructs with dnTCF4 shows that activation of the TOPflash reporter by Barx2 is inhibited by dnTCF4. D. Cotransfection of Barx2, constructs with MyoD shows that these factors synergize in regulation of TOPflash activity. E. Cotransfection of FL-Barx2 with MyoD, Myf5, MRF4, or myogenin shows that Barx2 can synergize with all four MRFs. F. Cotransfection with FL-Barx2 or the variant lacking the Eh-1 motif (Barx2ΔEh1), with or without full length (Grg-l) or truncated (Grg-s) Groucho isoforms in C2C12 cells shows that Groucho is involved in regulation of TOPflash; Grg-l blocks activation by Barx2ΔEh1 but not wildtype Barx2, while Grg-s blocks activation by both wildtype Barx2 and Barx2ΔEh1. Luiciferase activity was assayed 48 hours post-transfection; all data were normalized to a Renilla luciferase internal control and then to pcDNA3 transfection. Data were collected in triplicate and at least 2 assays were performed for each condition with similar results. In panels A, B, C, D, and F, ** and * indicate changes relative to the control (pcDNA) that are significant at P<0.01 and P<0.05 respectively. In panel E, ** and * indicate changes relative to the control (Barx2) that are significant at P<0.01 and P<0.05 respectively.