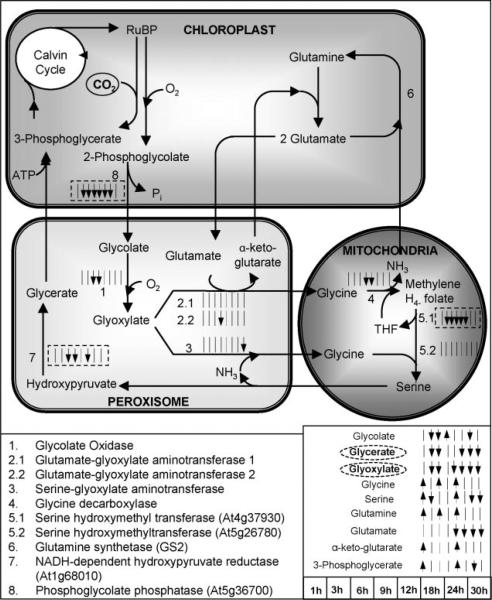
Figure 5.
The significance level profiles over time and the significance classification based on paired-SAM of the genes and metabolites in the photorespiration pathway that were included in the analysis. The transcriptional information is compartmentalized as shown in the figure, i.e., all shown genes are expressed in the particular compartments. The significance level profiles of metabolites that participate in this pathway are shown outside of the compartments, because they refer to the net over the entire liquid culture (i.e., over all compartments, tissue and cell types) metabolite pool size. If the gene encoding a particular reaction's enzyme was identified as positively or negatively significant at a time point (see time point sequence in the lower right side of the figure), the corresponding small time point arrow next to the reaction is facing up or facing down, respectively. If the gene was identified as non-significant at a time point, then a small straight line is shown in the respective location. A solid or dashed, respectively, box around the time point arrows signifies that the gene was identified as positively or negatively significant by paired-SAM. If a metabolite was identified as positively or negatively significant at a particular time point, the corresponding time point arrow next to this metabolite's name is facing up or down, respectively. If a metabolite was identified as positively or negatively significant by paired-SAM then its name is circled by a straight or dashed, respectively, line. Information about the photorespiration pathway structure was obtained from the KEGG database (www.kegg.com) and Buchanan et al. (2001). Dotted lines depict positive or negative regulation depending on the sign. (This figure can be viewed online in a color version as Supplementary Figure 1.)