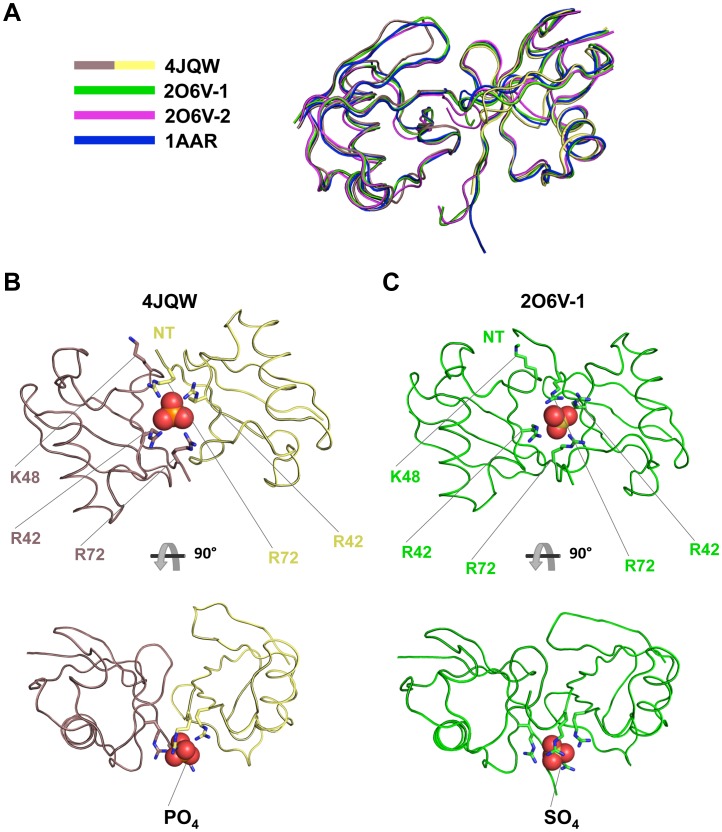
Figure 2. Mono-Ub in our structure formed dimers similar in conformation to other structures of K48-linked di-Ub.
A) Ribbon representation of an alignment of K48-linked diubiquitin structures. The proximal di-Ub (2O6V-1, green) and distal di-Ub (2O6V-2, magenta) from tetra-Ub and di-Ub from 1AAR (blue) were aligned to the Ub dimer structure reported in this study (Ub C, mauve taupe and Ub D, yellow). Main chain RMSDs for this alignment were 1.309, 0.892 and 0.737 Å, respectively. B) Loop representation of the Ub dimer from our structure shown chelating a phosphate anion. This chelation is mediated mainly by Arg42 and Arg72 from each subunit. C) Loop representation of the proximal di-Ub moiety from K48-linked tetra-Ub (2O6V-1). In this case, Arg42 and Arg72 mediate chelation of a sulfate anion. In both structures, the Arg residues, which are located at the periphery of the hydrophobic pocket and often participate in interactions with UBDs, are engaged instead through these chelation interactions. This may serve as a mechanism by which UBDs discriminate between K48-linked poly-Ub and other forms of poly-Ub. K48 of the distal Ub moiety and the N-terminus (NT) of the proximal Ub moiety of each dimer are shown, demonstrating the close proximity in the dimer relative to the covalently linked chain.