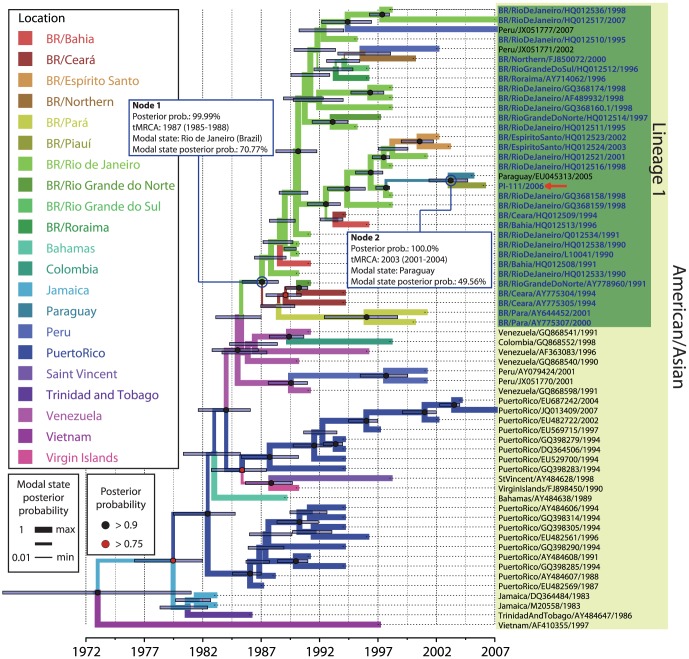
Figure 4. Bayesian coalescent and discrete phylogeographic analyses of Brazilian DENV-2 lineage 1 based on envelope nucleotide sequences.
A maximum clade credibility tree was inferred by Bayesian inference analysis (strict molecular clock; TN93+G; 200,000,000 iterations) using 63 DENV-2 envelope sequences (summarized in Table S1) retrieved by a BLAST search against the entire GenBank database and using PI-111/2006 (indicated by a red arrow) as a query.PI-111/2006 is an isolate from the state of Piauí (Brazil) that clustered in Brazilian DENV-2 lineage 1. Nodes that presented posterior probability value of >0.9 and >0.75 are represented by black and red circle ( ), respectively. Blue bars in each node represent the extent of the 95% highest probability density (95% HPD) for each divergence time. The most probable geographic state for each internal node was inferred by discrete phylogeographic analysis. Different colors in the branch represent distinct geographical states according to the legend on the left side of the figure. The branch width is proportional to the probability value of the inferred ancestral geographical state.
), respectively. Blue bars in each node represent the extent of the 95% highest probability density (95% HPD) for each divergence time. The most probable geographic state for each internal node was inferred by discrete phylogeographic analysis. Different colors in the branch represent distinct geographical states according to the legend on the left side of the figure. The branch width is proportional to the probability value of the inferred ancestral geographical state.