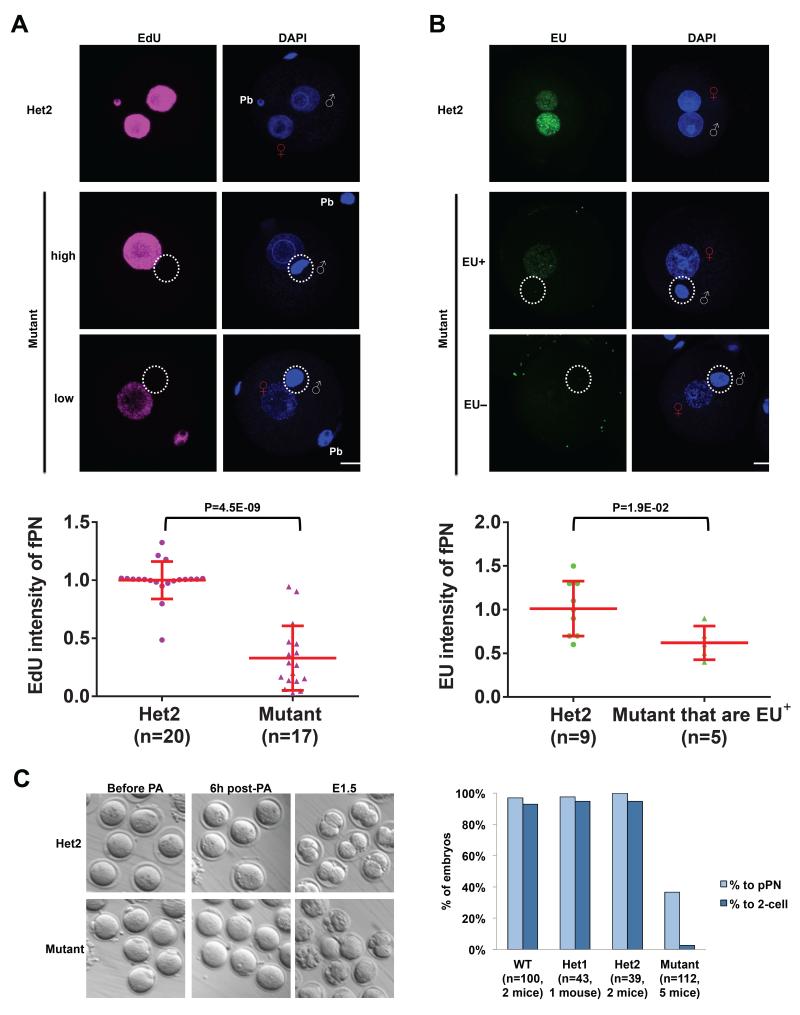
Figure 3. Maternal Hira regulates DNA replication and RNA transcription in both parental genomes.
(A) DNA replication as detected by EdU staining is consistently absent in the paternal genome, while it is reduced in the maternal genome in Hira mutant zygotes. Representative EdU labeling images in heterozygous zygotes and high- or low-EdU-intensity in Hira mutants are shown (upper panel). Quantification of EdU intensity of female pronucleus (fPN) in Hira mutant and heterozygous zygotes (lower panel).
(B) EU labeling indicates a complete loss of nascent RNA synthesis in the paternal genome and a severe deficiency in the maternal genome in Hira mutant zygotes (upper panel). 54% of mutant zygotes have maternal genomes with no detectable EU signal (hence quantification not shown), while the remaining 46% show significantly reduced levels of EU-positivity (quantified in the lower panel). Two-tailed t-test with unequal variance was used and error bars indicate s.d..
(C) Hira mutant oocytes fail to undergo cleavage to form 2-cell embryos after parthenogenetic activation (PA) (left panel). Many mutant oocytes show visible cellular fragmentation the day after PA, while almost all control oocytes formed a pseudo-pronucleus (pPN) and proceeded to the 2-cell stage (right panel). Scale bar: 20 m. White “♂” symbol or white dashed circle indicates male chromatin; Red “♀” symbol indicates female chromatin; Pb refers to a polar body.
See also Figure S3.