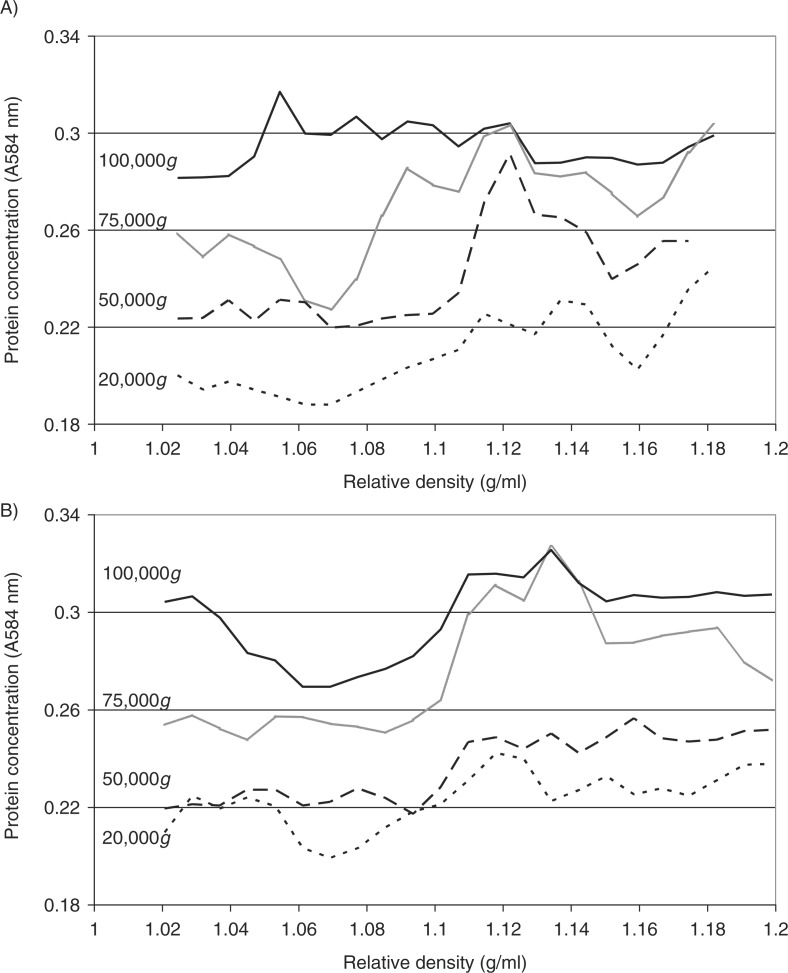
Fig. 3.
Analysis of the relative density of microvesicles by density gradient centrifugation. Cells lines (MDA-MB-231 and A375) were washed and pre-adapted to respective serum-free media prior to activation with PAR2-AP (SLIGRL; 20 µM). The conditioned media were collected and cleared of any cell debris by centrifuging at 5,400g for 10 min on a microcentrifuge. Cell-derived microvesicles were then sedimented at various RCF ranging from 20,000 to 100,000g at 20°C for 1 h. Microvesicles recovered at various centrifugal forces were analysed by density gradient ultracentrifugation using a sucrose–OptiPrep gradient covering an approximate range of 1.02–1.22 g/ml against 2 sets of DensityMarkerBeads as density markers. The samples and markers were centrifuged at 52,000g for 90 min at 20°C. The distribution of the protein content of the microvesicles derived from (A) MDA-MB-231 and (B) A375 cells was analysed using the Bradford assay, by measuring the absorption at 584 nm and subtracting the values obtained from a similar set without loading the microvesicles. Traces were obtained by calculating the moving-average values calculated from the data.