Abstract
Early-onset dystonia is associated with the deletion of one of a pair of glutamic acid residues (c.904_906delGAG/c.907_909delGAG; p.Glu302del/Glu303del; ΔE 302/303) near the carboxyl-terminus of torsinA, a member of the AAA+ protein family that localizes to the endoplasmic reticulum (ER) lumen and nuclear envelope (NE). This deletion commonly underlies early-onset DYT1 dystonia. While the role of the disease-causing mutation, torsinAΔE, has been established through genetic association studies, it is much less clear whether other rare human variants of torsinA are pathogenic. Two missense variations have been described in single patients; R288Q (c.863G>A; p.Arg288Gln; R288Q) identified in a patient with onset of severe generalized dystonia and myoclonus since infancy, and F205I (c.613T>A, p.Phe205Ile; F205I) in a psychiatric patient with late-onset focal dystonia. In this study, we have undertaken a series of analyses comparing the biochemical and cellular effects of these rare variants to torsinAΔE and wild-type (wt) torsinA in order to reveal whether there are common dysfunctional features. The results revealed that the variants, R288Q and F205I, are more similar in their properties to torsinAΔE protein than to torsinAwt. These findings provide functional evidence for the potential pathogenic nature of these rare sequence variants in the TOR1A gene, thus implicating these pathologies in the development of dystonia.
Keywords: torsinA, dystonia, DYT1, endoplasmic reticulum, protein secretion, ER stress
Introduction
Dystonia is characterized by involuntary muscle contractions, which cause sustained twisting and repetitive movements, as well as abnormal postures [Fahn, 1988]. Early onset DYT1 generalized dystonia is the most common and severe form of the hereditary dystonias with autosomal-dominant inheritance and a reduced penetrance of 30–40% [Bressman et al., 1989]. In most cases, DYT1 is caused by a heterozygous 3 bp deletion (ΔGAG) in exon 5 of the TOR1A gene (MIM# 605204) corresponding to loss of one of the two adjacent glutamic acid residues at positions 302 or 303 in torsinA, torsinAΔE (c.904_906delGAG/c.907_909delGAG; p.Glu302del/Glu303del) [Ozelius et al., 1997]. Additional sequence variants which change an amino acid (a.a.) in torsinA have been found in the heterozygous state in other patients with dystonia, but their role in causing the disease has not yet been determined. These include an 18 bp deletion (c.966_983del18; p.Val322_Tyr328delinsVal) in exon 5, encoding a.a. 323 – 328, identified in a patient presenting with atypical dystonia with myoclonic features [Leung et al., 2001]. The potential pathogenic nature of this mutation is in doubt, however, as this patient also had a mutation in the epsilon sarcoglycan gene responsible for myoclonus dystonia [Doheny et al., 2002]. An aspartic acid (D) 216 to histidine (H) (c.646G>C; p.Asp216His) change has been found to influence penetrance of the ΔGAG mutation, but is not independently associated with dystonia onset [Kock et al., 2006; Risch et al., 2007; Kamm et al., 2008]. A 4 bp deletion (c.934_937delAGAG; p.Arg312_Val313delinsPhefs) causing a frameshift truncation was identified in an anonymous control sample for whom no neurologic evaluation was available [Kabakci et al., 2004] and later also found in a patient with late onset myoclonic and dystonic features with signs of Parkinson’s disease [Ritz et al., 2009]. Other missense variants reported include an arginine (R) 288 to glutamine (Q) substitution (c.863G>A; p.Arg288Gln) in a patient with severe generalized dystonia [Zirn et al., 2008], and a phenylalanine (F) 205 to isoleucine (I) substitution (c.613T>A, p.Phe205Ile) in a psychiatric patient with late-onset focal dystonia [Calakos et al., 2010].
The TOR1A gene belongs to the family of AAA+ proteins (ATPase associated with variety of cellular activities) [Ozelius et al., 1997; Neuwald et al., 1999], most of which form hexameric rings in their active conformation, often assembled from inactive dimers [Beuron et al., 2003; DeLaBarre and Brunger, 2003; Krzywda et al., 2002]. The nucleotide binding sites of AAA+ proteins lie at the interface between subunits permitting determinants from adjacent subunits to contribute to nucleotide hydrolysis. Like other AAA+ proteins torsinA may be functional as multimers with torsinAΔE inhibiting the activity of wild-type (wt) torsinA [Kustedjo et al., 2000; Gordon and Gonzalez-Alegre, 2008; Breakefield et al., 2001; Torres et al., 2004; Goodchild et al., 2005; Hewett et al., 2007; Hewett et al., 2008; Chen et al., 2010a; Nery et al., 2011].
TorsinA is localized in the lumen of the endoplasmic reticulum (ER) and contiguous nuclear envelope (NE) of most cells, with torsinAΔE being more concentrated in the NE and, when overexpressed, producing whorled membrane inclusions derived from NE/ER membranes [Hewett et al., 2000; Hewett et al., 2003; Kustedjo et al., 2000; Gonzalez-Alegre and Paulson, 2004; Goodchild and Dauer, 2004], as does overexpression of a torsinB variant [Rose et al., 2014]. Although the function of torsinA is still unclear, a variety of properties in cultured cells change upon expression of torsinAΔE and/or down-regulation of torsinA levels consistent with a role in membrane-cytoskeleton interactions. In the ER, torsinA has been associated with processing of proteins through the secretory pathway [Torres et al., 2004; Hewett et al., 2007; Hewett et al., 2008], degradation of proteins by ER associated degradation (ERAD) [Esapa et al., 2007; Nery et al., 2011], the ER stress response [Chen et al., 2010a; Nery et al., 2011] and extension of neurites [Hewett et al., 2006; Nery et al., 2008; Atai et al., 2012]. In the NE, torsinA is involved in nuclear polarization during cell migration [Nery et al., 2008], morphology of NE membranes [Naismith et al., 2004; Goodchild et al., 2005], and nuclear egress of large ribonuclear protein (RNP) complexes [Jokhi et al., 2013] and herpes simplex virus (HSV) [Maric et al., 2011].
To better understand the impact of torsinA variants associated with dystonia (torsinAΔE, torsinAF205I and torsinAR288Q) on cellular phenotype, and the mechanism of action of torsinA, we used homology modeling approaches as well as biochemical and cell based assays to compare activities of torsinAwt with these variants. This study confirms that these torsinA variants have features similar to torsinAE and hence are implicated in causing dystonia.
Materials and Methods
Predicted Structural Features of TorsinA Variants
Bioinformatics sequence analysis with SignalP 4.1 [Petersen et al., 2011] revealed the nature of the first 60 a.a. residues of torsinA. Residues 1 to 20 were identified as a signal sequence (SS) and residues 21 to 60 were predicted by TMHMM [Moller et al., 2001] to reside in a membrane-associated domain. Hence, these residues were left out of the modeling procedure. We used the homology modeling approach, implemented in the MODELLER [Sali and Blundell, 1993] 9v10 program, to build models of torsinAwt, delE302, delE303, F205I and R288Q based on the 3D structure of the ClpB second AAA+ domain of Thermus thermophilus (T. thermophilus) [Lee et al., 2003]. The protocol used to perform the molecular modeling experiments was: generation of 10 models, from which one model for each torsinA sequence was selected. All models were submitted to the DOPE energy scoring function [Shen and Sali, 2006] implemented in the MODELLER 9v10 aiming to select the best structures. The MOLPROBITY webserver [Chen et al., 2010b] and PROCHECK [Laskowski et al., 1993] were used to verify and validate the stereochemical quality of the models. Intermolecular hydrogen bonds (HB) were calculated and displayed with the program LIGPLOT [Wallace et al., 1995]. All images were generated with the PyMOL program (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
Molecular Dynamics Simulations
Five independent molecular dynamic (MD) simulations and trajectory analyses were carried out with the GROMACS 4.5.5 package [Hess et al., 2008] for wt and variants of torsinA at room temperature or 298.15 K. Each dimeric model contained approximately 180,000 atoms, including ATP, explicit water molecules, and counter ions (Na+ and Cl−) to neutralize the system. The CHARMM27 force field [MacKerell et al., 1997] was used for proteins. Parameters for ATP and TIP3P water molecules were taken from the AMBER force field [Duan et al., 2003 and Jorgensen et al., 1983, respectively]. The LINCS algorithm [Hess et al., 1997] was used to constrain all covalent bonds allowing an integration time step of 2 fs. The systems’ temperature was slowly increased in five steps (50 K, 100 K, 200 K, 250 K, and 298.15 K), each lasting 200 ps. An equilibration phase (5 ns) was obtained without any position restraint within an NPT ensemble at 298.15 K and 1 atm. Each MD simulation lasted for 20.0 ns and average values were calculated for the last 12.0 ns of the MD trajectory during which the system is considered equilibrated.
DNA Constructs
Human torsinA cDNAs (NM_000113.2), 999 base pairs (bp), encoding torsinAwt and ΔE were generated from controls and DYT1 patients and introduced into EcoRI and NotI restriction sites of pcDNA3.1+ (Invitrogen Life Technologies, Darmstadt, GERMANY), generating pcDNA3-torsinAwt, pcDNA3-torsinAΔE (302/303) [Ozelius et al., 1997]. To generate the pcDNA3-torsinAR288Q construct, the Expand Long Template PCR System (Roche Applied Science, Mannheim, GERMANY) was used to introduce the R288Q mutation into the pcDNA3-torsinAwt construct using the primer, 5‘-GTGGAAATGCAGTCCCAAGGCTATGAAATTGATG-3‘ and 5‘-CATCAATTTCATAGCCTTGGGACTGCATTTCCAC-3‘ with a subsequent DpnI digestion. To generate the pcDNA-torsinAF205I construct, the Quickchange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) was used to introduce the F205I mutation into the pcDNAwt construct using the primers, 5′-CCAGAAAGCCATGTTCATAATTCTCAGCAATGCTGGAGC-3′ and 5′-GCTCCAGCATTGCTGAGAATTATGAACATGGCTTTCTGG-3′, as described previously [Calakos et al., 2010].
All cDNAs encoding human torsinA variants were also cloned into the NheI and XhoI sites of the CSCW2-IRES lentivirus vector plasmid [Shu et al., 2006]. CSCW2 is a self-inactivating lentiviral vector, which has a cytomegalovirus (CMV) immediate early promoter controlling transgene expression with the monomeric Cherry (mCherry) fluorescent protein encoded after the internal ribosomal entry site (IRES) element [Sena-Esteves et al., 2004]. All final vectors were sequenced to confirm sequence identities. Lentiviral constructs expressing secreted Gaussia luciferase (Gluc) and firefly luciferase (Fluc), CSCW2-Gluc-IRES-GFP [Badr et al., 2007] and CSCW2-Fluc-IRES-mCherry [Maguire et al., 2008] were prepared, as described [Sena-Esteves et al., 2004].
Statistical Analysis
Data were analyzed using two-tailed Student’s t test (Excel, Microsoft, Redmond, WA, USA) or factorial analysis of variance (ANOVA) as applicable using InStat v3.0 (GraphPad Software, La Jolla, CA, USA). After detection of a statistically significant difference in a given series of treatments by ANOVA, post-hoc Dunnett’s t-tests were performed when appropriate. The p-values <0.05 were considered statistically significant (shown as a single asterisk in the figures); p-values <0.01 were considered highly statistically significant (shown as a double asterisk in the figures), and p-values <0.001 (shown as triple asterisk in the figures).
Results
Predicted Structural Features of Variant Forms of TorsinA
The crystal structure of T. thermophilus ClpB has been described previously [Lee et al., 2003] and was used as a template to generate a structural framework for torsinA [Kock et al., 2006]. Human torsinA presents 332 a.a., which can be divided into four regions. The first 20 a.a. are predicted to be a SS. Residues 21 to 60 are mostly hydrophobic and predicted to be a membrane-associated domain (Fig. 1A). Therefore, no acceptable Protein Data Bank (PDB) templates were found for these regions of the protein. The other two additional domains, the AAA+ domain (L61 to N246) and the C-terminal domain (N247 to D332), were fully modeled in this work. The positions of variant sequences found in torsinA in dystonia patients are indicated in Figure 1A. In the case of the torsinAΔE deletion, it is not clear whether a.a. E302 or E303 is deleted. Other sequence variations in patients with dystonic symptoms include missense variations in exon 3 (F205I; [Calakos et al., 2010]) and exon 5 (R288Q; [Zirn et al., 2008]).
Figure 1. Predicted structural features of torsinA variants, their dimerization and molecular dynamics.
(A) Overall organization and sequence variations in torsinA (SS = signal sequence; Hypb = hydrophobic domain; AAA+ domain; and C-terminal domain). (B) Overall, structure of human torsinA. AAA+ domain is composed by 11 alpha helices (light green) and 5 beta strands (dark green), and the C-terminal domain is composed by 3 alpha helices, where the alpha 13 and 14 are colored in light blue and alpha 15 is in dark blue. ATP molecule is represented by its chemical structure (orange, purple and yellow). (C) Location of each of the four torsinA mutations studied is superimposed on the torsinAwt model. In torsinAwt a.a. carbons are colored in orange and the mutants are colored in yellow. Image generated with PyMol. (D) Lysates of BE(2)C cells (30 µg) expressing torsinAwt or torsinA variants: torsinAΔE, torsinAR288Q, and torsinAF205I were electrophoresed under non-reducing and reducing (200 mM DTT) conditions and gels immunoblotted with antibodies to torsinA or beta-actin. (E) Quantification of the band intensity of 75 kDa "dimers" in non-reducing conditions for torsinAwt, torsinAΔE, torsinAR288Q, and torsinAF205I relative to levels of beta-actin (ANOVA post-hoc Dunnett's t-test *p<0.05; **p<0.01; ***p<0.001; n=5). (F) Molecular dynamics analyses of torsinAwt and variants.
The ClpB and torsinA alignment, previously proposed [Kock et al., 2006] was used as a reference for modeling torsinAwt (Fig. 1B), and to analyze the consequences of four sequence variants (delE302, delE303, F205I and R288Q) in the torsinA structure (Fig. 1C). The models for torsinAwt, torsinAF205I, and torsinAR288Q results in a 273 a.a. chain while the torsinAΔE models (delE302 and delE303) have 272 a.a. each. As proposed in Kock et al. [2006], the human torsinA topology is similar to that of the C-terminal region of ClpB proteins. The human torsinA models proposed herein depict fourteen α-helices, α1 (Q62-N68), α2 (H73-V83), α3 (K108-Y120), α4 (F132-A134), α5 (F138-I144), α6 (Q152-L153), α7 (N158-C162), α8 (G178-L187), α9 (A209-L219), α10 (L240-V242), α11 (N246-N249), α12 (K275-Y290), α13 (E303-V313) and α14 (T321-TY329) and five β-strands, β1 (L98-H101), β2 (N127-L129), β3 (I166-N170), β4 (M202-S207) and β5 (P268-L270). With regard to the organization of these structural elements, this protein has a large domain composed of a five strand β-sheet surrounded by eleven α-helices, and a smaller domain made of three α-helices [Zhu et al., 2008]. Figure 1B shows a three-dimensional (3-D) model of human torsinA.
It has been shown that proteins having the same or very similar sequences may have significantly different structures (Kosloff and Kolodny, 2008). Our studies show that the overall structures of the AAA+ and C-terminal domains of torsinAwt compared to torsinAΔE (delE302 and delE303), torsinAF205I and torsinAR288Q do not indicate major changes in conformation resulting from the sequence variation (Fig. 1B). All models possess both well-characterized domains. However, we observed the present of some differences in a number of secondary structural elements, notably the helices. All torsinA secondary structures have a different number of α-helices and in the small region linking the three helices of the C-terminal domain. Mutation F205I is located in the AAA+ domain, specifically at strand β4. Based on our homology modeling, this mutation does not result in any major or local structural changes when compared with wt. The other mutations are all located in the C-terminal domain. The R288Q mutation lies within subdomain α5. This subdomain is adjacent to subdomain α6 that is affected by the ΔE deletion. Further, removing E302 may not have a large impact on secondary structure because it is at the border between Helix 15 (Supp. Figure S1). Conversely, when we remove E303, we remove a residue found within Helix 15 (these measurements use torsinAwt as a reference). Insertions and deletions are usually well accommodated when they occur at turns and loops connecting secondary structural elements. The different structures with identical sequences are used as starting conformations for the MD simulations. As observed from our homology modeling we have found some secondary structural changes in the new torsinA variants, and they may play roles in oligomer assembly and/or interaction with torsinA’s partners compromising its function. We further investigated these possibilities by subjecting the models to simulation using the MDs method, as described below.
Analyses of TorsinA Interactions (putative dimers)
Similar to other AAA+ proteins, torsinA is predicted to assemble into oligomers [Breakefield et al., 2001]. Self-association is a general and conserved feature of members of the AAA+ family and changes in oligomerization have been correlated with changes in function [Mogk et al., 2003]. A 75 kDa torsinA immunoreactive band, in addition to the predicted monomeric 37 kDa torsinA band, has been observed by SDS-PAGE under non-reducing conditions for torsinAΔE [Torres et al., 2004; Gordon and Gonzalez-Alegre, 2008], consistent with the formation of dimer complexes. We asked whether this oligomerization of torsinA is also affected by the other two sequence variants found in dystonia patients, F205I and R288Q. Lysates from BE(2)C cells transduced with lentivirus vectors expressing torsinAwt, torsinAΔE, torsinAR288Q, or torsinAF205I were exposed to non-reducing and reducing conditions, and proteins resolved by SDS gel electrophoresis with immunoblotting for torsinA. While all three torsinA sequence variants resulted in both the 37 kDa form, as well as a stable higher molecular weight (MW) species of 75 kDa under non-reducing conditions, torsinAwt was primarily present as the 37 kDa species with lower tendency to form the 75 kDa species. The higher MW form is predicted to be torsinA dimers and disappears under reducing conditions (Fig. 1D). Although the intensity of the 75 kDa band varied between experiments, it was consistently found with torsinA variants and only rarely and weakly with torsinAwt (Fig. 1E), demonstrating a clear biochemical difference associated with the torsinA variants. These results suggest that dimerization in cells is enhanced by these torsinA variants.
Molecular Dynamics Simulations for TorsinA
Computational MDs simulations have a growing role in the understanding of the behavior of a variety of macromolecular systems and their interactions [van Gunsteren and Mark, 1998]. These include analysis of molecular complex formation and stability [Monecke et al., 2013; Norberto de Souza and Ornstein, 1999], predictions of enzyme-substrate activity [Chien et al., 2012], effects of mutations on drug resistance [Schroeder et al., 2005], and inhibitor binding [Costa et al., 2012].
To relate the putative dimerization results with torsinA expression in neuroblastoma cells, MD simulations (in aqueous solution in the presence of counter ions (Na+ and Cl−) to simulate a physiologic buffer) were performed to monitor the stability of dimers of torsinAwt and torsinA variants (delE302 and delE303, F205I and R288Q), as well as the affinity of these models for ATP. Figure 1F summarizes the results of radius of gyration (RG) and HB pattern with the ATP ligand. RG is a measure of protein structure compactness. The lower the RG the tighter the packing of the protein while higher RG values indicate loosely packed proteins (Flory, 1953). As depicted in Figure 1F, torsinAwt and the delE302 present higher deviations in solution when compared with the variants delE303, R288Q, F205I, and the reference RG value (19.0 ± 0.1 Å) for proteins of the same class and size as torsinA [Lobanov et al., 2008]. Since RG is a measure of the compactness of torsinA dimers, these results suggest that torsinAwt and delE302 may form looser dimers, relative to the other variants. This is in agreement with our SDS-PAGE/western blot analysis, which showed that the torsinA variants have an increased tendency to form dimers (Fig. 1D–E).
The HB pattern of the ATP-torsinA interaction along the MD simulation trajectory was also determined. The average values of HB formation for each model are shown in Figure 1F. Modeling revealed that the mutants have fewer HBs coupled to ATP relative to torsinAwt. Kock et al. [2006] also proposed that torsinA variants could be interfering with ATP association. F205I and delE303 models have the lowest number of HBs, compared to other variants and torsinAwt. Overall, the modeling and MD simulation results support and, at least in part, explain some of the experimental data in this work, as well as implicating E303 as the mutation underlying DYT1 dystonia.
Furthermore, lentiviral vectors encoding Fluc (for normalization) and secreted Gluc were used to transduce BE(2)-C neuroblastoma cells previously transduced with lentiviral vectors expressing either torsinAwt or torsinA variants, in the configuration CMV-torsinA-IRES-mCherry (Fig. 2A). Three days after transfection, neuroblastoma BE(2)-C cells expressing torsinAwt or torsinA disease variants and mCherry were selected by fluorescence-activated cell sorting (FACS) (Supp. Figure S2, see Supp. Materials and Methods). These sorted populations expressed mCherry at similar levels. The recovered cell lines maintained stable expression over 10 passages in culture.
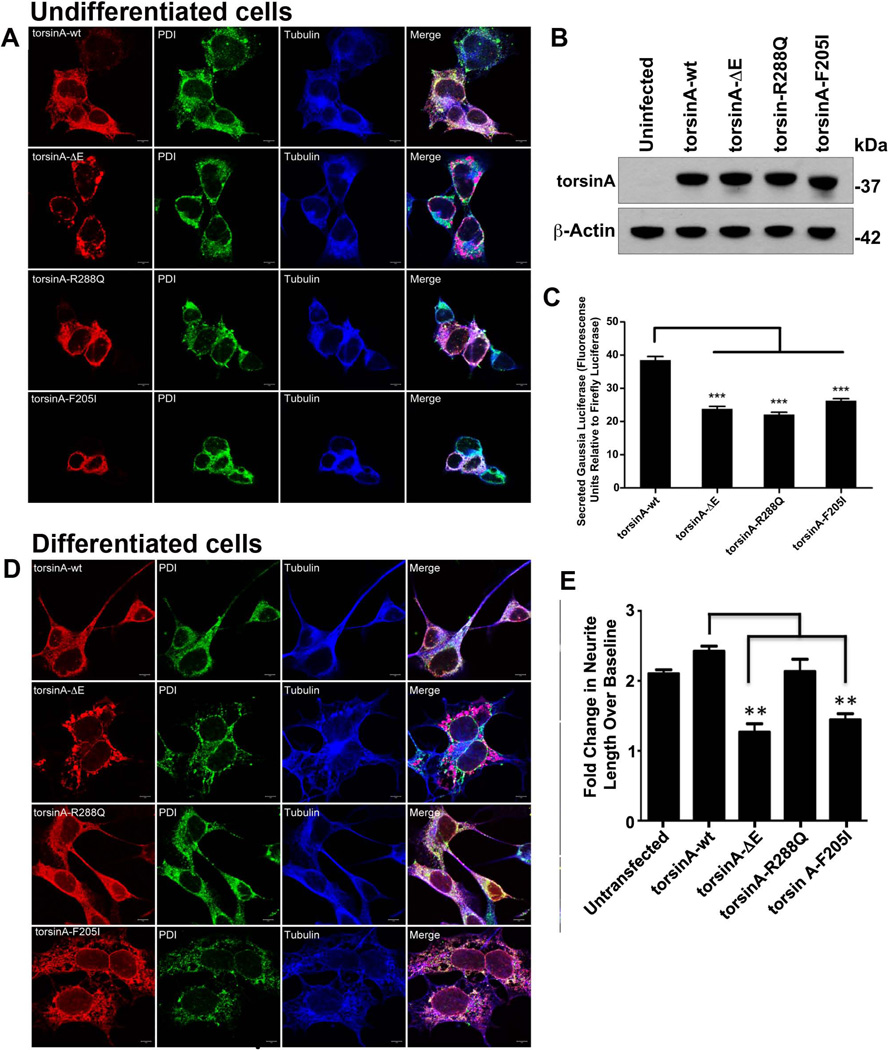
Figure 2. BE(2)C cells expressing variant forms of torsinA presented decreased Gluc secretion and neurite outgrowth as compared with cell expressing torsinAwt.
(A) BE(2)C cells were transduced with the lentiviral vectors encoding torsinAwt, torsinAΔE, torsinAR288Q or torsinAF205I. Forty-eight hr later cells were immunostained for torsinA, PDI, and β-tubulin. (B) BE(2)C cells non-transduced or transduced with vectors expressing wt or torsinA variants were lysed and 30 µg of each sample resolved by SDS PAGE. A representative western-blot shows equal levels of torsinA in all transduced cells. (C) BE(2)C cells expressing torsinA variants were transduced with lentiviral vectors encoding Gluc-IRES-cerulean and Fluc-IRES-mCherry. The cells were replated 48 hr after infection at 6 × 104 cells per well in 96-well plates. The Gluc activity in the medium was determined as RLU at 24 hr after replating the cells. (D) BE(2)C stable cell lines expressing torsinA forms: torsinAwt torsinAΔE, torsinAR288Q, and torsinAF205I were grown for 3 days in presence of blebbistatin and all-trans retinoic acid and visualized by immunocytochemistry using a polyclonal torsinA antibody TA2 (red), a monoclonal antibody specific for PDI (green), and monoclonal antibody β-tubulin-Alexa Fluor 647 (blue). (E) Measurement of neurite length after treatment with blebbistatin and retinoic acid for 3 days in uninfected BE(2)C cells and cells infected with lentiviral vectors encoding torsinAwt and torsinA variants. A two-fold increase corresponds to growth of about 40 µm. The experiments were repeated three times in quadruplicates for each cell line (ANOVA post-hoc Dunnett's t-test ***p<0.001; n=3).
These cells show the typical ER association of torsinA as indicated by co-localization with the ER chaperone protein, PDI, as well as the tendency of torsinAΔE to form ER-derived inclusions [Hewett et al., 2003]. Cell lysates resolved by SDS-PAGE with immunostaining for torsinA and β-actin showed similar levels of expression of torsinA in transduced cells regardless of the variant expressed (Fig. 2B). Non-infected cells have low endogenous expression of torsinA, which was not detected here due to the high dilution of antibody used to monitor overexpression.
Using these cells, we assessed Gluc flux through the secretory pathway by measuring luciferase activity in the medium over time normalized to the Fluc activity in cells (Fig. 2C). Neuroblastoma cells expressing torsinA variants showed decreased levels of Gluc activity in the medium relative to those expressing torsinAwt (Fig. 2C), indicating a common impairment of protein secretion in cells expressing these torsinA variants, as previously found for torsinAΔE in other cell types [Hewett et al., 2007; Hewett et al., 2008].
In order to determine whether expression of torsinAR288Q and torsinAF205I altered neurite length, we differentiated transduced BE(2)-C human neuroblastoma cells and compared these with non-transduced cells. The cell morphology (Fig. 2D) and length of neurites (Fig. 2E) were analyzed three days post-differentiation. While cells transduced with lentiviral vectors expressing torsinAwt and torsinAR288Q showed similar morphologies, expression of torsinAΔE and F205I resulted in gross abnormalities in both ER and microtubule structure (Fig. 2D). This is consistent with previous reports that torsinAΔE forms inclusions derived from the ER and suggests that torsinAF205I may lead to ER vacuolization. Moreover, no difference in the neurite number was observed between cells expressing torsinA variants and torsinAwt. While no changes in the final neurite length was observed between non-transduced BE(2)-C cells or cells expressing either torsinAwt or torsinAR288Q, corresponding to an increase in neurite length of 40 ± 7 µm in 3 days. BE(2)-C cells expressing torsinAΔE and torsinAF205I showed significantly shorter neurites of 28 ± 6 µm after the same time period (Fig. 2E).
Characterization of VSVG Transport Under Conditions of TorsinA Knock-down
To further elucidate a potential role for torsinA in protein processing, we traced the movement of YFP-tagged vesicular stomatitis virus glycoprotein (VSVG)ts045 through the secretory pathway. By regulating environmental temperature, unfolded VSVG can be loaded into the ER and then visualized as it traffics to the Golgi complex, where the protein temporarily accumulates while undergoing N-glycosylation. Trafficking continues onto the plasma membrane (PM), where the protein ultimately localizes. To determine the role of endogenous torsinAwt in this process, an adenovirus vector expressing YFP-VSVGts045 [Keller et al., 2001] was used to transduce 293T cells with and without silencing of torsinA using si1958 or si1963 (Fig. 3A–B; see Supp. Materials and Methods). VSVG trafficking from the ER to the Golgi complex was visualized by confocal microscopy after torsinA depletion relative to scrambled siRNA transduced cells (Fig. 3A; see Supp. Materials and Methods). Quantification of the kinetics of VSVG trafficking from the ER to the Golgi complex over a 10 min period showed a delay in transport to the Golgi when torsinAwt levels were reduced (Fig. 3C). Blinded image analysis was used to quantitate the percent of VSVG in the ER (Fig. 3D) and Golgi complex (Fig. 3E) over 20 min. Down-regulation of torsinA significantly increased the amount of VSVG retained in the ER by 3.2-fold at 10 min and 10.9-fold at 20 min, with a corresponding decrease of about 50% in the Golgi complex at 20 min.
Figure 3. Absence of torsinA or overexpression of variant forms of torsinA impairs ER exit of YFP-VSVG.
293 T cells transfected with torsinA siRNA (si1958 and si1963) or scrambled siRNA as a control were infected with adenovirus expressing YFP-tagged VSVGts045 (VSVG) and incubated at 40.5°C for 16 hr. Following this incubation, cells were either fixed immediately (time 0 - ER loaded VSVG), or transferred to a 32°C water bath and trafficking of VSVG was assessed. (A) In scrambled control cells, ER loaded VSVG (0 min) can be visualized (green fluorescence) as it traffics in vesicles (10 min, arrow heads) to the Golgi complex (20 min) where the protein temporarily accumulates while undergoing N-glycosylation (20 min, arrow heads). Trafficking of VSVG from the ER to the Golgi complex showed a defect in ER exit following loss of torsinA (arrows). (B) Knock-down efficiency was confirmed after 72 hr by western analysis. Quantification of ER associated VSVG shows a decrease in rate of VSVG exit from the ER following torsinA silencing after 10 min (C) and 20 min (D). (E) Quantification of Golgi complex-associated VSVG shows a decrease in rate of Golgi complex accumulation following torsinA silencing (n=3). In addition, uninfected 293T cells, and 293T cells expressing torsinAwt, and variant forms: torsinAΔE, torsinAR288Q and torsinAF205I were infected with adenovirus expressing YFP-tagged VSVGts045 (VSVG) and incubated, as described above. (F) Representative fluorescent images of cells stained for torsinA (red) and YFP-VSVG (green). (G) Image analysis quantification of the ER localization of VSVG in cells expressing different torsinA isoforms after 10 min; and (H) after 20 min at 32°C. Expression of variant torsinA forms are associated with delayed exit of VSVG from the ER in 293T cells after 10 and 20 min. Quantification of Golgi complex-associated VSVG shows a decrease in the rate of Golgi complex accumulation in torsinA variants (n=4). (Scale bars = 10 µm, ANOVA post-hoc Dunnett's t-test *p<0.05, **p<0.01).
In addition, we used an antibody specific to the folded form of VSVG [Lefrancois and Lyles, 1982] to evaluate whether this protein was being folded properly in the ER under conditions of torsinA depletion (Supp. Figure S3). The appearance of folded VSVG protein was evident in 74% of cells treated with scrambled siRNA within 1 min of allowing cells to recover from temperature induced blockade of protein processing (Supp. Figure S3A). Under conditions of torsinA depletion (cells treated with siRNAs 1958 and 1963; Supp. Figure S3B, C, and D), however, VSVG folded protein was largely absent until 5 min post-recovery, at which time only 29% of cells were immuno-positive for folded VSVG (Supp. Figure S3E). This supports the role of torsinA as a chaperone in the processing of this secreted protein.
VSVG Transport in Presence of Wild-type and Variant Forms of TorsinA
VSVG trafficking was also evaluated in 293T cells stably transduced with lentivirus vectors expressing equivalent amounts of torsinAwt, torsinAΔE, torsinAR288Q, or torsinAF205I. VSVG localization in cells was monitored over 20 min by immunocytochemistry and fluorescence microscopy following a shift to the permissive temperature (Fig. 3F). Image analysis revealed that exit from the ER was almost twice as high in cells expressing torsinAwt, as compared to expression of all the other variants at 10 min (Fig. 3G). This trend continued out to 20 min with torsinAR288Q apparently being the most competent among the torsinA variants in facilitating the trafficking of VSVG cargo (Fig. 3H). The differences in the VSVG trafficking were not related to differential loading of VSGV in the ER, as we observed comparable levels of VSVG in the ER after 16 hr at 40.5°C, the restrictive temperature, in all cells independent of the type of torsinA expressed (Supp. Figure S4).
Protein Exit from the ER is Not Affected by the Presence of Wild-type and Variant Forms of TorsinA
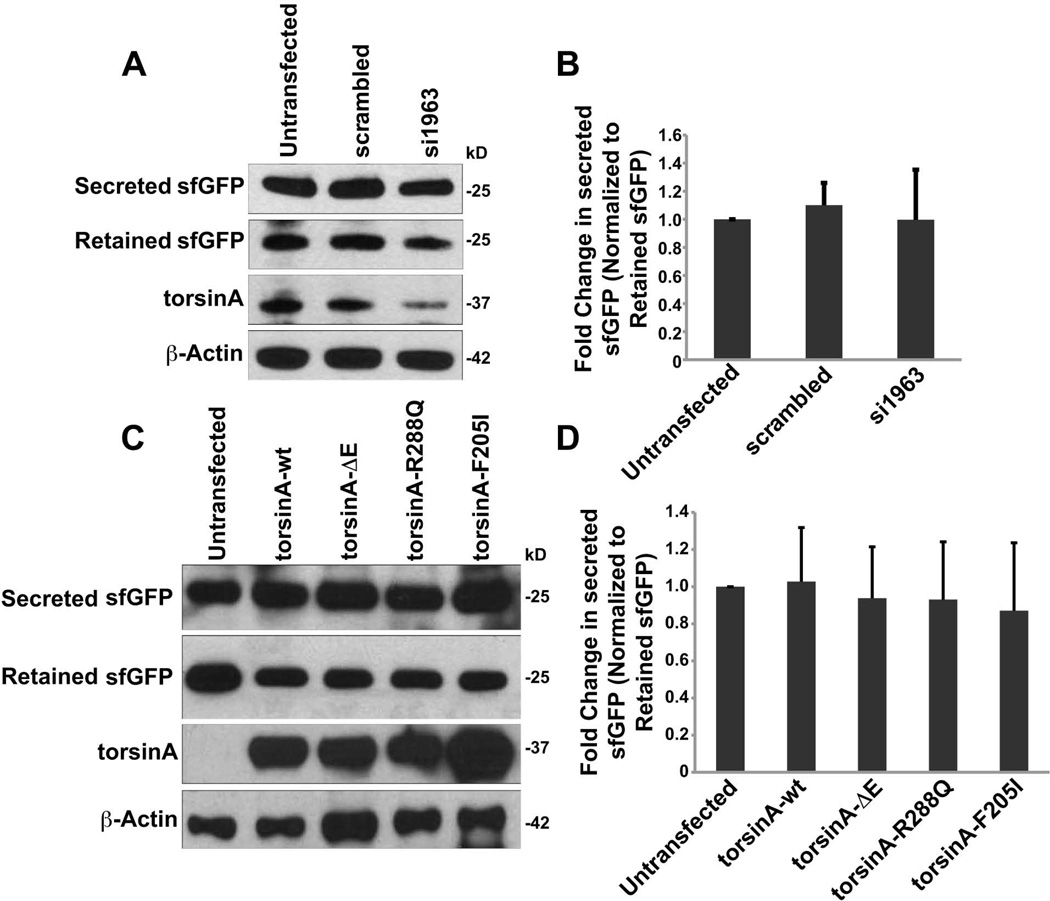
To test the role of torsinA in the exit of proteins from the ER, 293T cells expressing torsinAwt and variant forms: torsinAΔE, torsinAR288Q and torsinAF205I were transfected with an expression cassette encoding a sec-sfGFP as a soluble cargo [Aronson et al., 2011; Pédelacq et al., 2006]. Sec-sfGFP is known to possess robust folding properties that allow it to exit the ER through the secretory pathway without assistance from ER chaperone proteins. To evaluate the extent of GFP secreted in the medium, we normalized expression to the amount retained in cells. In order to inhibit expression of torsinA, siRNA oligonucleotides were used to knock-down torsinAwt mRNA. siRNAs were transfected into 293T cells and torsinA and actin levels were measured 72 hr later by western blot. siRNA 1963 was able to selectively decrease torsinA levels, but did not change the amount of sec-sfGFP secreted or retained (Fig. 4A). Quantitative densitometry of sec-sfGFP showed no difference in the levels of sec-sfGFP secretion or retention in torsinA-silenced versus siRNA scrambled controls when normalized to α-actin levels (Fig. 4B). In addition, we measured sec-sfGFP secretion in cells transduced with lentivirus vectors encoding torsinAwt and torsinA variants (torsinAΔE, torsinAR288Q, and torsinAF205I) (Fig. 4C). Neither overexpression of torsinAwt or torsinA mutant/variants had any effect on the extent of sec-sfGFP secretion (Fig. 4D), indicating that torsinA is not involved in the regulation of ER exit sites.
Figure 4. TorsinA role in the secretion of sfGFP.
The expression of torsinA variants and torsinA knock-down did not affect the secreted levels of sfGFP (n=3). (A and B) TorsinAwt in 293T cells was down-regulated by siRNA for three days followed by transfection with sfGFP for 10 hr. siRNA reduced torsinAwt levels by >50%, yet levels of sec-sfGFP remained unaltered relative to levels of retained sfGFP. (C and D) 293T cells expressing torsinAwt or torsinA variants (n=3) also did not display an effect on sec-sfGFP levels relative to retained sfGFP levels, suggesting that torsinA does not function in the regulation of ER exit sites in the secretory pathway.
Morphological Changes Caused by TorsinA Variants in Fibroblasts
Human fibroblasts were used to further evaluate ER morphology as their large cytoplasmic volume allows for ease of assessment of ER structure. Fibroblasts from a control individual were transduced with lentiviral vectors expressing torsinAwt or torsinA variants and then immunostained 72 hr later for torsinA and the ER protein, PDI (Fig. 5A). Western blot analysis confirmed similar expression levels of torsinA among lines (data not shown). Consistent with our findings in differentiated BE(2)-C cells, expression of torsinA R288Q in fibroblasts did not result in notable morphological changes in the ER, relative to cells expressing torsinAwt. A notable change was observed, however, in cells expressing either torsinAΔE or torsinAF205I, with torsinAE causing inclusions and the torsinAF205I form causing apparent ER vacuolization (Fig. 5A, lower panel in left). These findings are consistent with those found in BE(2)-C cells, with the apparent vacuolization also seen upon expression of a truncated form of torsinA (313–332) in 293T or fibroblasts [Nery et al., 2011]. The majority of the cells expressing the F205I construct showed evidence of ER vacuolization (Supp. Figure S5).
Figure 5. Expression of torsinA isoforms perturbs the morphology of the ER and NE.
Fibroblast cells infected with lentivirus expressing torsinAwt and torsinA variants. (A) Forty-eight hr post-infection cells were processed for immunocytochemistry for torsinA and PDI. Magnification bar = 10 µM. (B) NE ultrastructure in torsinAwt and torsinA isoforms expressing human fibroblasts cells. Cells expressing torsinAwt (nucleus on the left, cytoplasm on the right; top panel), and torsinA isoforms: NE in torsinAE, torsinAR288Q, torsinAF205I (in order from the top to the bottom). Note, abnormal spacing between inner and out nuclear membrane in cells expressing torsinA variants (scale bar = 100 nm). Higher magnification (scale bar = 500 nm) image shows abnormal spacing in torsinAR288Q (Fig. 5C) and torsinAFF205I (Fig. 5D) cell lines. Note, the herniation of inner nuclear membrane into perinuclear space in cells (Fig. 5C–D).
We next examined the ultrastructure of the NE by electron microscopy (EM; see Supp. Materials and Methods). To confirm that the abnormalities in nuclear membrane structure were not a result of torsinA staining, we did not label torsinA in our EM studies. In fibroblasts transduced with the torsinAwt construct, EM analysis showed a uniform spacing between the outer and inner nuclear membranes typical of NE ultrastructure (Fig. 2B). By contrast, focally enlarged perinuclear spaces, and ballooning into large bubbles were found in the NE of cells overexpressing torsinAΔE, torsinAR288Q or torsinAF205I (compare Fig. 5B upper panel with three bottom panels). Expression of torsinAR288Q and torsinAF20I showed sites where the inner nuclear membrane was separated from the nuclear lamina, producing membrane-delimited herniations into the perinuclear space (Fig. 5C–D).
Discussion
These studies employ a number of assays which have previously demonstrated marked differences between torsinAwt and torsinAΔE when overexpressed in cultured cells, including a tendency to dimerize in the absence of reducing agents, reduced processing of Gluc and VSVG through the secretory pathway, decreased neurite extension and alterations in ER/NE morphology. These assays were used to evaluate the consequences of defects in torsinA function in two missense variant forms of torsinA, torsinAR288Q and torsinAF205I, found in patients with dystonia. As with torsinAΔE, both these torsinA variants showed an increased tendency to dimerize, reduced processing of Gluc and VSVG, and altered NE morphology. The R288Q mutation appeared to have less impact on normal torsinA function relative to the F205I mutation, as only the latter showed compromised neurite extension and grossly altered ER morphology. Since the R288Q mutation is associated with severe generalized early onset dystonia [Zirn et al., 2008], and the F205I mutation with late onset, focal dystonia [Calakos et al., 2010], this suggests that torsinA properties of dimerization, protein processing and NE morphology may contribute to the pathophysiology of dystonia. Collectively, these data provides a means of assessing the impact of coding sequence variations in torsinA on cellular pathogenicity, as well as providing a series of tests to measure potential efficacy of novel therapeutic agents.
AAA+ proteins, such as torsinA, typically form six-membered homo-oligomeric rings that control both assembly and disassembly of numerous protein complexes involved in protein processing, membrane trafficking, cytoskeletal dynamics and organelle biogenesis [Vale, 2000; Ogura and Wilkinson, 2001; Hanson and Whiteheart, 2005]. The closest orthologue of torsinA with known 3D structure is the second AAA+ domain of the bacterial heat-shock protein ClpB, derived from T. thermophilus [Lee et al., 2003; Iyer et al., 2004]. To assess the effects of missense mutations or deletions within the AAA+ or C-terminal domain of torsinA on its function, we built structural homology models for torsinAwt, F205I, R288Q, delE302, and delE303 (Fig. 1B and 1C) using the 3D structure of ClpB as a template. Given the close topological relationship between torsinA and the ClpB template [Kock et al., 2006], we did not expect much structural variation from these static models. We therefore performed MD simulations for the dimers of all models generated in an explicit aqueous solution. In MD simulations macromolecular systems may relax over the period of simulation time, allowing them to accommodate to intrinsic and environmental alterations. The changes we observed in RG during MD simulation suggests that the dimer models of torsinAwt and delE302 are less compact relative to those for F205I, R288Q, and delE303. The looser dimers observed in models of torsinAwt and delE302 may be due to a weaker association between their respective monomers, while the higher compactness predicted in models for F205I, R288Q, and delE303 may by contrast indicate stronger associations. This hypothesis is supported by western blotting experiments that indicated a greater tendency of torsinA variants to form dimers under non-reducing conditions. Interestingly, HB analysis of torsinA-ATP interaction models showed that torsinAwt has a higher affinity for ATP than torsinA disease variants. While R288Q is located in the C-terminal domain, F205I is found in strand β4 closer to the ATP binding site in the AAA+ domain (Fig. 1C). The proximity of F205I seems to result in long range structural changes that may affect its ability to bind ATP. The differences in HB pattern (Fig. 1F) and the relative positions of R288Q and F205I mutations may explain why F205I has a more prominent impact on torsinA function in biological assays. Deletion of E303 predicts a severe decrease in torsinA binding to ATP (6 HBs) when compared to either wt (13 HBs) or deletion of E302 (10 HBs). These results are consistent with those of Kock et al. [2006] and Zhao et al. [2013] in predicting which torsinA variants interfere with ATP binding affinity. In addition, the B-factor behavior showed that deletion E303 is more structurally deleterious than E302. We can rationalize this finding based on the changes in helix α6 of the C-terminal domain. According to the torsinA/ClpB pairwise sequence alignment (Fig. 1B of 8) residues E302 and E303 are the first two residues of helix α6. Residue E302 is at the boundary with the loop connecting helix α5 to α6. It is highly flexible and its conformation may transition between coil and helical conformations. Residue E303 is located towards the interior of helix α6, and therefore its deletion may cause local structural disorders that can propagate towards both the N- and C-terminus of the helix. Hence, deletion E303 is more disturbing to the structure. The spatial long range effects of these changes results in poor packing of the C-terminal three-helix bundle domain. Although this structural disorder enhances the probability of delE303 dimerization, as observed through the RG parameter behavior, it is not compensated by the very low affinity for ATP.
Previous studies have suggested that differences in oligomerization of torsinA can influence its rate of degradation [Torres et al., 2004; Gordon and Gonzalez-Alegre, 2008]. Self-association is a general and conserved feature of members of the AAA+ family of ATPases and can influence AAA+ protein function [Mogk et al., 2003; Oguchi et al., 2012]. Thus, we examined whether torsinA self-associates and whether this property was altered in any of the torsinA variants. All mutants showed an increased tendency to dimerize in cell lysates under non-reducing conditions with torsinAΔE>R228Q>F205I>wt. This was consistent with structural predictions and may indicate a more stable structure of mutant oligomers relative to wt oligomers. This may suggest a dominant-negative effect of heterozygotic variants in patient cells with mutant forms presumably binding to the wt forms and thereby reducing their activity.
Our previous studies demonstrated that trafficking of proteins through the secretory pathway can be monitored using a naturally secreted, and highly sensitive luciferase, Gluc [Tannous et al., 2005] and that DYT1 patient cells have a marked decrease in the rate of Gluc secretion as compared with control cells [Hewett et al., 2007]. This appeared to be due to reduced function of torsinA as an ER chaperone protein, as mouse embryonic fibroblasts (MEFs) from homozygous torsinA knock-out mice also showed reduced Gluc secretion, relative to wt MEFs [Hewett et al., 2007]. In addition, knock-down experiments using siRNAs that reduced levels of wt torsinA decreased Gluc secretion, while reduced levels of torsinAΔE increased secretion in DYT1 fibroblasts [Hewett et al., 2008]. Here, we expanded these findings by monitoring trafficking of YFP-VSVGts045 from the ER to the Golgi complex. Decreased levels of torsinA following siRNA knock-down of torsinA markedly reduced this trafficking (by 80%), again implicating torsinA in protein processing in the ER. Since sec-sfGFP exit from the ER was not impaired, delayed trafficking of YFP-VSVGts045 from the ER to the Golgi complex is likely due to impaired protein processing in the ER. This is consistent with our finding that torsinA depletion impairs normal folding of YFP-VSVGts045 in the ER and with studies showing that impaired processing of proteins in cells expressing endogenous or elevated levels of torsinAΔE show ER stress, presumably due to protein overload [Hewett et al., 2003; Nery et al., 2011; Chen et al., 2010a].
Further, in previous studies using human neuroblastoma SH-SY5Y cells we showed the overexpression of torsinAΔE delays the kinetics of neurite outgrowth relative to torsinAwt [Hewett et al., 2006]. In the present study we find that overexpression of the torsinA variant F205I, similarly compromises neurite extension in human neuroblastoma cells. While F205I results are consistent with those of torsinAΔE, a more detailed morphological analysis is needed to ascertain the basis of these defects in neurite growth. Abnormalities in brain structure have been reported in mouse models with altered torsinA expression, including delayed interkinetic nuclear migration during neurogenesis and impaired migration of neurons in torsinA homozygous knock-out embryos [McCarthy et al., 2012] and changes in size and dendrite morphology of neurons in heterozygous torsinA knock-in mice [Song et al., 2013]. Reduction in neuronal fiber tracts have also been observed in the brains of heterozygous DYT1 knock-in mice [Uluğ et al., 2011].
Overexpression of all three torsinA mutant variants described herein resulted in abnormal spacing between the inner and outer nuclear membranes, including presence of intraluminal vesicles. These findings are consistent with those reported following torsinAΔE overexpression in a variety of cell types, as well as absence torsinA in mouse homozygous knock-out embryos [Gonzalez-Alegre and Paulson, 2004; Naismith et al., 2004; Goodchild et al., 2005]. This is also consistent with the defined role of torsinA in modulating nuclear membrane dynamics. This function is dependent on torsinA's interaction with nesprins, which span the outer nuclear membrane and anchor the NE to the cytoskeleton [Ketema and Sonnenberg, 2011]. TorsinA also appears to mediate association between SUN proteins in the inner NE and nesprins in the outer NE, as a component of the LINC complex [Vander Heyden et al., 2011; Jungwirth et al., 2011]. The consequences of mutations in or down-regulation of torsinA in the NE include restricted movement of the nucleus, manifesting as a delay in cell migration [Hewett et al., 2008], and the nuclear export of both large ribonuclear particles [Jokhi et al., 2013] and HSV-1 virions [Maric et al., 2011].
Mutation or depletion of torsinA impacts a variety of cellular processes, including nuclear polarization, cell migration, export of ribonuclear particles from the nucleus, protein folding, neurite extension and the maintenance of NE and ER morphology. The ability of torsinA to simultaneously impact so many discrete cellular compartments may be due to association with multiple proteins that span the ER and NE membranes and their interaction with cytoskeletal elements. Our results showing that torsinA variants form more stable dimers may suggest differences in their ability association numerous proteins, as compared to torsinAwt, thus impacting on a multitude of cellular functions. It may thus seem surprising that so few mutations have been identified in torsinA that result in dystonia. Mutations in torsinA that result in a null allele in combination with a wt allele are fully compatible with normal motor function, while complete loss of expression, as seen in homozygous knock-out mice, results in neonatal death [Goodchild et al., 2005; Dang et al., 2005]. The mutations causing dystonia in the heterozygous state must therefore be unique and exert a dominant-negative effect that blocks normal torsinA activity. Structural analysis of the disease-causing torsinA mutants reported in this paper appears to compromise the region of the protein predicted to bind to ATP. This region also appears to affect dynamic aspects of oligomerization with the mutants forming more stable complexes with each other and possibly torsinAwt. Alternatively, they may impede binding to other proteins, such as LULL1 associated with ATPase activity [Zhao et al., 2013]. Collectively, the assays used in the present study provide a means both to evaluate the role of specific torsinA mutants in the disease process, as well as to validate efficacy of therapeutics in correcting or overriding the dominant-negative effects of mutant torsinA. These assays implicate both torsinAR228Q and torsinAF205I as additional mutations causing autosomal dominant dystonia in humans.
Supplementary Material
Acknowledgments
We thank Ms. Suzanne McDavitt for skilled editorial assistance. We thank Dr. Nicole Calakos, Dr. Naoto Ito, and Dr. Noriko Wakabayashi-Ito for helping with torsinA F205I construct. siRNAs for torsinA mRNA and scrambled siRNA were kindly provided by Alnylam Inc. through Dr. Dinah Sah.
Grant support: This work was supported by NIH/NINDS grant NS037409 (XOB, FCN), Lentivirus vectors were prepared by the NINDS funded Neuroscience MGH Vector Core, Boston, MA, supported by NIH/NINDS P30NS045776 (XOB, BAT). Flow core NIH grant support 1S10RR023440-01A1 (FN). ONS thanks the National Research Council of Brazil (CNPq) for grants 305984/2012-8 and 559917/2010-4 and CAPES for a Ph.D. scholarship to LFSMT. SDR was supported by a fellowship from the Canadian Institute of Health Research and the Dystonia Medical Research Foundation. KG was supported by the Deutsche Forschungsgemeinschaft, the Elitepostdoc Programme Baden Württemberg, Germany, and the IZKF University of Tuebingen.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Aronson DE, Costantini LM, Snapp EL. Superfolder GFP is fluorescent in oxidizing environments when targeted via the Sec translocon. Traffic. 2011;12:543–548. doi: 10.1111/j.1600-0854.2011.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atai NA, Ryan SD, Kothary R, Breakefield XO, Nery FC. Untethering the nuclear envelope and cytoskeleton: biologically distinct dystonias arising from a common cellular dysfunction. Int'l J Cell Biology. 2012;2012:634214. doi: 10.1155/2012/634214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2:e571. doi: 10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS. Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. J Mol Biol. 2003;327:619–629. doi: 10.1016/s0022-2836(03)00178-5. [DOI] [PubMed] [Google Scholar]
- Breakefield XO, Kamm C, Hanson PI. TorsinA: movement at many levels. Neuron. 2001;31:9–12. doi: 10.1016/s0896-6273(01)00350-6. [DOI] [PubMed] [Google Scholar]
- Bressman SB, de Leon D, Brin MF, Risch N, Burke RE, Greene PE, Shale H, Fahn S. Idiopathic dystonia among Ashkenazi Jews: evidence for autosomal dominant inheritance. Ann Neurol. 1989;26:612–620. doi: 10.1002/ana.410260505. [DOI] [PubMed] [Google Scholar]
- Calakos N, Patel V, Gottron M, Wang G, Tran-Viet KN, Brewington D, Beyer JL, Steffens DC, Krishnan RR, Zuchner S. Functional evidence implicating a novel TOR1A mutation in idiopathic, late-onset focal dystonia. J Med Genet. 2010;47:646–650. doi: 10.1136/jmg.2009.072082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Burdette AJ, Porter CJ, Ricketts JC, Fox SA, Hewett JW, Nery FC, Berkowitz LA, Breakefield XO, Caldwell KA, Caldwell GA. The early-onset torsion dystonia associated protein, torsinA, is a homeostatic regulator of endoplasmic reticulum stress response. Hum Mol Genet. 2010a;19:3502–3515. doi: 10.1093/hmg/ddq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010b;D66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CH, Gao QZ, Cooper AJ, Lyu JH, Sheu SY. Structural insights into the catalytic active site and activity of human Nit2/ω-amidase: kinetic assay and molecular dynamics simulation. J Biol Chem. 2012;287:25715–25726. doi: 10.1074/jbc.M111.259119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa ALP, Pauli I, Dorn M, Schroeder EK, Zhan C-G, Norberto de Souza O. Conformational changes in 2-trans-enoyl-ACP (CoA) reductase (InhA) from M. tuberculosis induced by an inorganic complex: a molecular dynamics simulation study. J Mol Model. 2012;18:1779–1790. doi: 10.1007/s00894-011-1200-7. [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, McNaught KS, Jengelley TA, Jackson T, Li J, Li Y. Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp Neurol. 2005;196:452–463. doi: 10.1016/j.expneurol.2005.08.025. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- Doheny D, Danisi F, Smith C, Morrison C, Velickovic M, De Leon D, Bressman SB, Leung J, Ozelius L, Klein C, Breakefield XO, Brin MF, et al. Clinical findings of a myoclonus-dystonia family with two distinct mutations. Neurology. 2002;59:1244–1246. doi: 10.1212/wnl.59.8.1244. [DOI] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Esapa CT, Waite A, Locke M, Benson MA, Kraus M, McIlhinney RA, Sillitoe RV, Beesley PW, Blake DJ. SGCE missense mutations that cause myoclonus-dystonia syndrome impair epsilon-sarcoglycan trafficking to the plasma membrane: modulation by ubiquitination and torsinA. Hum Mol Genet. 2007;16:327–342. doi: 10.1093/hmg/ddl472. [DOI] [PubMed] [Google Scholar]
- Fahn S. Concept and classification of dystonia. Adv Neurol. 1988;50:1–8. [PubMed] [Google Scholar]
- Flory PJ. Principles of Polymer Chemistry. First ed. Ithica, NY: Cornell University Press; 1953. [Google Scholar]
- Gonzalez-Alegre P, Paulson HL. Aberrant cellular behavior of mutant torsinA implicates nuclear envelope dysfunction in DYT1 dystonia. J Neurosci. 2004;24:2593–2601. doi: 10.1523/JNEUROSCI.4461-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: an effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci U S A. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Gordon KL, Gonzalez-Alegre P. Consequences of the DYT1 mutation on torsinA oligomerization and degradation. Neurosci. 2008;157:588–595. doi: 10.1016/j.neuroscience.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JG. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comp. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Ziefer P, Li S, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO. Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum Mol Genet. 2000;9:1403–1413. doi: 10.1093/hmg/9.9.1403. [DOI] [PubMed] [Google Scholar]
- Hewett J, Ziefer P, Bergeron D, Naismith T, Boston H, Slater D, Wilbur J, Schuback D, Kamm C, Smith N, Camp S, Ozelius LJ, et al. TorsinA in PC12 cells: localization in the endoplasmic reticulum and response to stress. J Neurosci Res. 2003;72:158–168. doi: 10.1002/jnr.10567. [DOI] [PubMed] [Google Scholar]
- Hewett JW, Nery FC, Niland B, Ge P, Tan P, Hadwiger P, Tannous BA, Sah DW, Breakefield XO. siRNA knock-down of mutant torsinA restores processing through secretory pathway in DYT1 dystonia cells. Hum Mol Genet. 2008;17:1436–1445. doi: 10.1093/hmg/ddn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett JW, Tannous B, Niland BP, Nery FC, Breakefield XO. Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc Natl Acad Sci U S A. 2007;104:7271–7276. doi: 10.1073/pnas.0701185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett JW, Zeng J, Niland BP, Bragg DC, Breakefield XO. Dystonia-causing mutant torsinA inhibits cell adhesion and neurite extension through interference with cytoskeletal dynamics. Neurobiol Dis. 2006;22:98–111. doi: 10.1016/j.nbd.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAAfl ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Morre MJ, Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3:988–995. doi: 10.1016/j.celrep.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Physics. 1983;79:926–935. [Google Scholar]
- Jungwirth MT, Kumar D, Jeong DY, Goodchild RE. The nuclear envelope localization of DYT1 dystonia torsinA-ΔE requires the SUN1 LINC complex component. BMC Cell Biol. 2011;12:24. doi: 10.1186/1471-2121-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabakci K, Hedrich K, Leung JC, Mitterer M, Vieregge P, Lencer R, Hagenah J, Garrels J, Witt K, Klostermann F, Svetel M, Friedman J, et al. Mutations in DYT1: extension of the phenotypic and mutational spectrum. Neurology. 2004;62:395–400. doi: 10.1212/01.wnl.0000113024.84178.f7. [DOI] [PubMed] [Google Scholar]
- Kamm C, Fischer H, Garavaglia B, Kullmann S, Sharma M, Shrader C, Grundmann K, Klein C, Borggraefe I, Lobsien E, Kupsch A, Nardocci N, et al. Susceptibility to DYT1 dystonia in European patients is modified by the D216H polymorphism. Neurology. 2008;70:2261–2262. doi: 10.1212/01.wnl.0000313838.05734.8a. [DOI] [PubMed] [Google Scholar]
- Keller P, Toomre D, Diaz E, White J, Simons K. Multicolour imaging of post-Golgi sorting and trafficking in live cells. Nat Cell Biol. 2001;3:140–149. doi: 10.1038/35055042. [DOI] [PubMed] [Google Scholar]
- Ketema M, Sonnenberg A. Nesprin-3: a versatile connector between the nucleus and the cytoskeleton. Biochem Soc Trans. 2011;39:1719–1724. doi: 10.1042/BST20110669. [DOI] [PubMed] [Google Scholar]
- Kock N, Naismith TV, Boston HE, Ozelius LJ, Corey DP, Breakefield XO, Hanson PI. Effects of genetic variations in the dystonia protein torsinA: identification of polymorphism at residue 216 as protein modifier. Hum Mol Genet. 2006;15:1355–1364. doi: 10.1093/hmg/ddl055. [DOI] [PubMed] [Google Scholar]
- Kosloff M, Kolodny R. Sequence-similar, structure-dissimilar protein pairs in the PDB. Proteins. 2008;71:891–902. doi: 10.1002/prot.21770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywda S, Brzozowski AM, Verma C, Karata K, Ogura T, Wilkinson AJ. The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution. Structure. 2002;10:1073–1083. doi: 10.1016/s0969-2126(02)00806-7. [DOI] [PubMed] [Google Scholar]
- Kustedjo K, Bracey MH, Cravatt BF. TorsinA and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275:680–685. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Lefrancois L, Lyles DS. Interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology. 1982;121:157–167. [PubMed] [Google Scholar]
- Leung JC, Klein C, Friedman J, Vieregge P, Jacobs H, Doheny D, Kamm C, DeLeon D, Pramstaller PP, Penney JB, Eisengart M, Jankovic J, et al. Novel mutation in the TOR1A (DYT1) gene in atypical early onset dystonia and polymorphisms in dystonia and early onset parkinsonism. Neurogenetics. 2001;3:133–143. doi: 10.1007/s100480100111. [DOI] [PubMed] [Google Scholar]
- Lobanov MY, Bogatyreva NS, Galzitskaya OV. Molecular Biology. Pleiades Publishing, Inc; 2008. Radius of gyration as an indicator of protein structure compactness; pp. 623–628. [Google Scholar]
- MacKerell AD, Bashford D, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Comput Chem. 1997;18:1463–1472. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, Pugh ENJ, Mingozzi F, Bennicelli J, Banfi S, Marshall KA, Testa F, Surace EM, Rossi S, Lyubarsky A, et al. Safety and efficacy of gene transfer for Leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric M, Shao J, Ryan RJ, Wong CS, Gonzalez-Alegre P, Roller RJ. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J Virol. 2011;85:9667–9679. doi: 10.1128/JVI.05314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DM, Gioioso V, Zhang X, Sharma N, Bhide PG. Neurogenesis and neuronal migration in the forebrain of the torsinA knockout mouse embryo. Dev Neurosci. 2012;34:366–378. doi: 10.1159/000342260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Schlieker C, Strub C, Rist W, Weibezahn J, Bukau B. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Monecke T, Haselbach D, Voß B, Russek A, Neumann P, Thomson E, Hurt E, Zachariae U, Stark H, Grubmüller H, Dickmanns A, Ficner R. Structural basis for cooperativity of CRM1 export complex formation. Proc Natl Acad Sci U S A. 2013;110:960–965. doi: 10.1073/pnas.1215214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc Natl Acad Sci U S A. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Armata IA, Farley JE, Cho JA, Yaqub U, Chen P, da Hora CC, Wang Q, Tagaya M, Klein C, Tannous B, Caldwell KA, et al. TorsinA participates in endoplasmic reticulum-associated degradation. Nat Commun. 2011;2:393. doi: 10.1038/ncomms1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO. TorsinA binds the KASH-domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genomic Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Norberto de Souza O, Ornstein RL. Molecular dynamics simulations of a protein-protein dimer: particle-mesh Ewald electrostatic model yields far superior results to standard cutoff model. J Biomol Struct Dyn. 1999;16:1205–1218. doi: 10.1080/07391102.1999.10508328. [DOI] [PubMed] [Google Scholar]
- Oguchi Y, Kummer E, Seyffer F, Berynskyy M, Anstett B, Zahn R, Wade RC, Mogk A, Bukau B. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat Struct Mol Biol. 2012;19:1338–1346. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de Leon D, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Risch NJ, Bressman SB, Sinthil G, Ozelius LJ. Intragenic cis and trans modification of genetic susceptibility in DYT1 torsion dystonia. Am J Hum Genet. 2007;80:1188–1193. doi: 10.1086/518427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz K, Gerrits MC, Foncke EM, van Ruissen F, van der Linden C, Vergouwen MD, Bloem BR, Vandenberghe W, Crols R, Speelman JD, Baas F, Tijssen MA. Myoclonus-dystonia: clinical and genetic evaluation of a large cohort. J Neurol Neurosurg Psychiatry. 2009;80:653–658. doi: 10.1136/jnnp.2008.162099. [DOI] [PubMed] [Google Scholar]
- Rose AE, Zhao C, Turner EM, Steyer AM, Schlieker C. Arresting a torsin ATPase reshapes the endoplasmic reticulum. J Biol Chem. 2014;289:552–564. doi: 10.1074/jbc.M113.515791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Schroeder EK, Basso LA, Santos DS, Norberto de Souza O. Molecular dynamics simulation studies of the wild-type, I21V and I16T mutants of isoniazid resistant Mycobacterium tuberculosis enoyl reductase (InhA) in complex with NADH: Towards the understanding of NADH-InhA different affinities. Biophys J. 2005;89:876–884. doi: 10.1529/biophysj.104.053512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Shen M-Y, Sali A. Statistical potential for assessment and prediction of protein structures. Prot Science. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- Song CH, Bernhard D, Hess EJ, Jinnah HA. Subtle microstructural changes of the cerebellum in a knock-in mouse model of DYT1 dystonia. Neurobiol Dis. 2013;62C:372–380. doi: 10.1016/j.nbd.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11:435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Torres GE, Sweeney AL, Beaulieu JM, Shashidharan P, Caron MG. Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc Natl Acad Sci U S A. 2004;101:15650–15655. doi: 10.1073/pnas.0308088101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluğ AM, Vo A, Argyelan M, Tanabe L, Schiffer WK, Dewey S, Dauer WT, Eidelberg D. Cerebellothalamocortical pathway abnormalities in torsinA DYT1 knock-in mice. Proc Natl Acad Sci U S A. 2011;108:6638–6643. doi: 10.1073/pnas.1016445108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gunsteren WF, Mark AE. Validation of molecular dynamics simulation. J Chem Physics. 1998;108:6109–6116. [Google Scholar]
- Vander Heyden AB, Naismith TV, Snapp EL, Hanson PI. Static retention of the lumenal monotopic membrane protein torsinA in the endoplasmic reticulum. EMBO J. 2011;30:3217–3231. doi: 10.1038/emboj.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Zhao C, Brown RS, Chase AR, Eisele MR, Schlieker C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc Natl Acad Sci U S A. 2013;110:E1545–E1554. doi: 10.1073/pnas.1300676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Wrabl JO, Hayashi AP, Rose LS, Thomas PJ. The torsin-family AAA+ protein OOC-5 contains a critical disulfide adjacent to Sensor-II that couples redox state to nucleotide binding. Mol Biol Cell. 2008;19:3599–3612. doi: 10.1091/mbc.E08-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirn B, Grundmann K, Huppke P, Puthenparampil J, Wolburg H, Riess O, Muller U. Novel TOR1A mutation p.Arg288Gln in early-onset dystonia (DYT 1) J Neurol Neurosurg Psychiatry. 2008;79:1327–1330. doi: 10.1136/jnnp.2008.148270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.