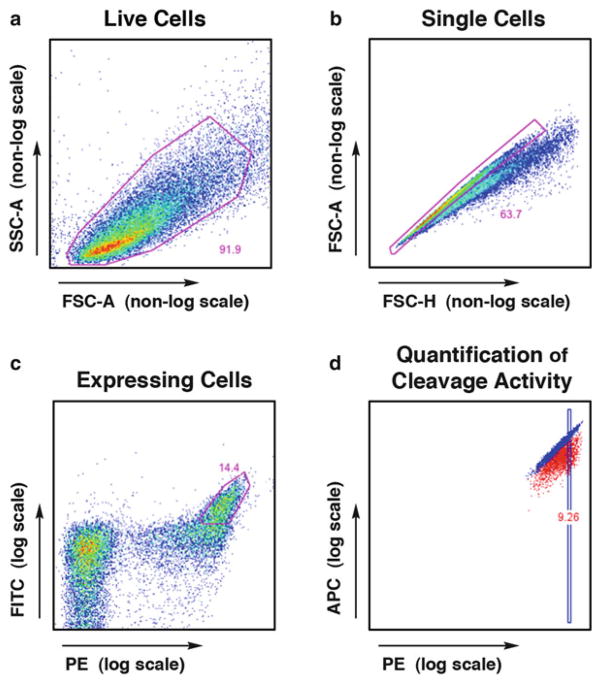
Fig. 7.
FlowJo analysis of flow cytometry data from the tethered DNA cleavage assay. (a ) Live cells are gated first, using FSC-A and SSC-A parameters, followed by (b) singlet cells using FSC-A and FSC-H. With proper induction conditions, approximately 50 % of cells will display the enzyme, and (c) the high-expressing population is gated using the PE signal (bound via a biotin–streptavidin bridge to the N-terminal HA tag) and FITC signal (bound directly by a fluorescence-conjugated antibody to the C-terminal Myc tag). Cleavage activity is visualized within this expressing population using a plot of (d) A647 fluorescence (observed in the APC channel, conjugated to the free end of the cleaved DNA) versus PE fluo-rescence (N-terminal HA tag). Cleavage activity can be quantified by gating on a PE-normalized subset of the expressing cells and calculating the ratio of median APC fluorescence for the uncleaved (Ca2+) versus cleaved (Mg2+) conditions