Figure 5. HK binds to apoptotic cells and is required for uPAR-mediated internalization.
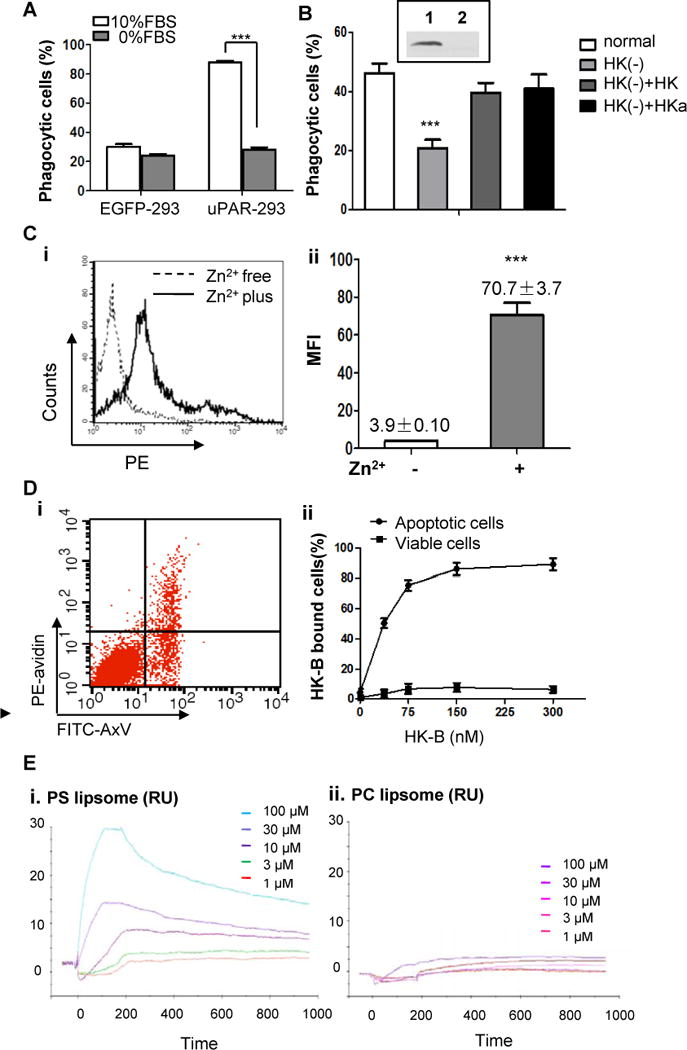
(A) EGFP-293 cells and uPAR-293 cells were co-cultured with apoptotic cells in the presence 10% FBS or 0% FBS for 2 hours. Internalization of apoptotic cells was analyzed by flow cytometry after treatment with TB as described in the legend for Figure 4. ***, p <0.001.(B) The levels of HK in normal serum (Lane 1, insert) and HK-depleted serum (lane 2, insert) were analyzed by immunoblotting. The uPAR-293 cells were incubated with apoptotic cells in presence of normal serum, HK-depleted serum [HK(−)], HK-depleted serum replenished with 50 nM HK [HK(−)+HK] or HKa [HK(−)+HKa], respectively. After treatment with TB, the internalization of apoptotic cells was evaluated by flow cytometry as described in the legend for Figure 4. ***, p< 0.001. (C) Apoptotic cells were incubated with 100 nM biotinylated HK (HK-B) in PBS with or without 50 μM ZnCl2 at 4°C for 15 minutes. After washing with PBS containing 0.35% BSA and 50 μM ZnCl2, the cells were stained with PE-avidin and the fluorescence intensity of was quantitated by flow cytometry. Representative data of HK bound to apoptotic cells in the presence (Zn2+ plus) or absence (Zn2+ free) of zinc ions (i) and the mean fluorescence intensity (MFI) from triplicate samples (ii) are shown. ***, p <0.001. (D) Mixed apoptotic cells and viable cells were incubated with 50 nM HK-B in Tyrode’s buffer containing 0.35% BSA and 50 μM ZnCl2 at 4°C for 15 minutes. After washing with PBS containing 50 μM ZnCl2, the cells were labeled with PE-avidin and non-saturated amounts of FITC-annexin V (AxV). The intensity of PE fluorescence for HK-B bound to apoptotic cells (FITC-AxV-positive) and viable cells (FITC-AxV-negative) was analyzed by flow cytometry (i). Shown in (ii) was the quantification of the percentage of PE-positive cells at the indicated concentrations of HK-B, representing HK-B bound cells (%). (E) SPR assay. PS liposome (i) or PC liposome (ii) at the indicated concentrations was allowed to bind to HK-immobilized CM5 sensor chip to reach equilibrium, followed by dissociation. The response curves were analyzed using BIAevaluation software.
