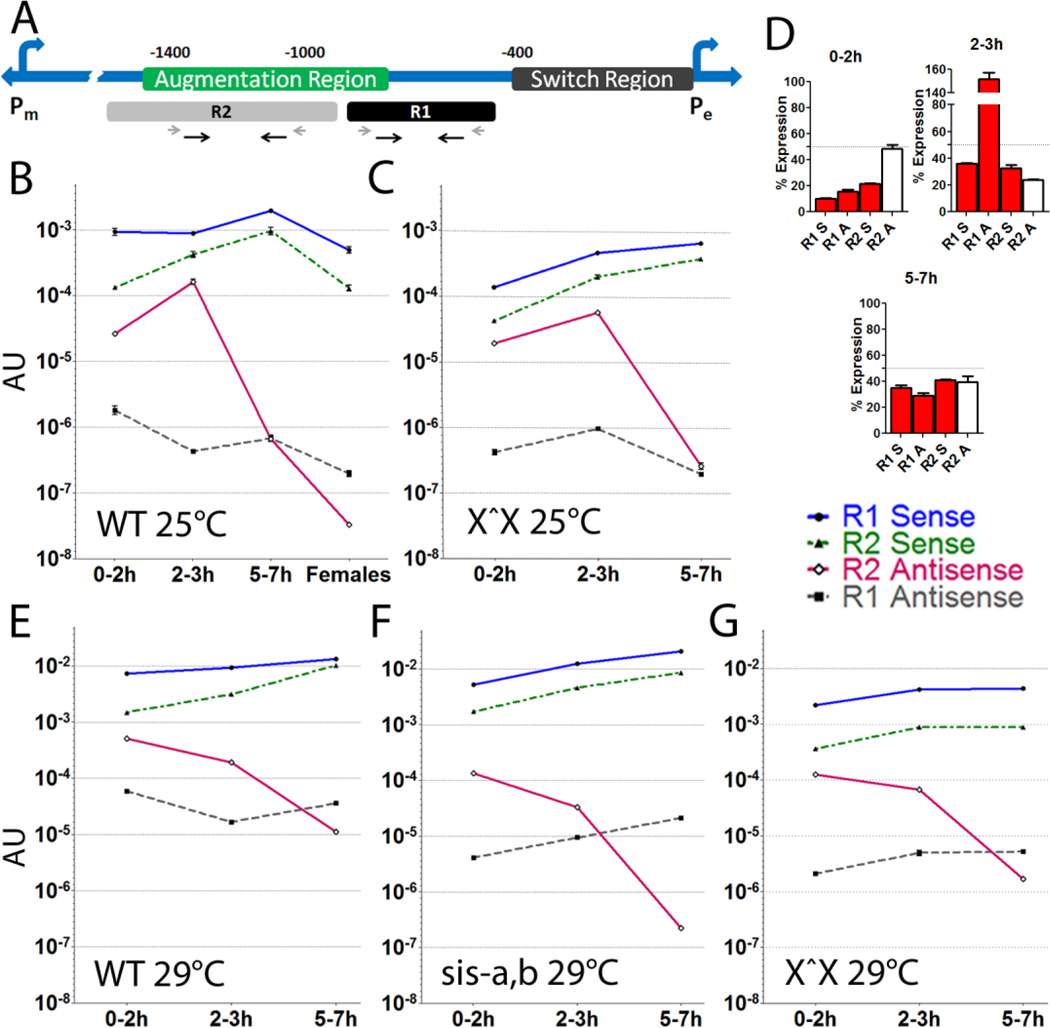
Figure 1. Drosophila sex determination utilizes lncRNAs near SxlPe.
(A) Positions of R1 and R2 lncRNA regions relative to known regulatory elements. The two Sxl promoters are indicated, SxlPm is ~5 kb upstream of SxlPe. Prior deletion studies of SxlPe [26] define the switch region as necessary for sex specific activation (0–0.4 kb upstream), while the augmentation region (0.8–1.4 kb upstream) is required for robust expression. Primers for PCR of R1 and R2 shown as black arrows, RT primers as shorter grey arrows. (B–G) Developmental profiles of the four R1 and R2 lncRNAs strands in different genotypes (key to strands on right). All measured by qRT-PCR and normalized to tubulin to give arbitrary units (AU) of expression. They are also corrected for approximate DNA content at each stage, to give a gauge of the lncRNA to DNA level (removing this correction does not significantly change the plot shapes, it mostly depresses the apparent 0–2h levels). Note the graphs are in log scale, which makes the small error bars difficult to see. (B) wild-type (OreR) 0–2, 2–3, 5–7h embryos and adult females. Expression of the 4 lncRNA strands relative to SxlPe mRNA (2–3 hr) is R1A: 0.000017, R1S: 0.03, R2A: 0.0057, R2S: 0.015. (C) attached-X stock where the free X chromosome has a deletion of Sxl (X^X; SxlfP7B0/Y). This allows scoring the female only signal. (D) Percent female AU for each lncRNA in 0–2, 2–3 and 5–7h embryos from an attached-X stock (data in C) relative to its corresponding lncRNA in wild-type (data in A), to give an indication of the proportion from males. The dotted line at 50% represents equal contribution from males and females. (E–G) Temperature and reduction of X chromosome counting genes depresses relative R2 antisense levels. (E) wild-type, (F) X^X SxlfP7B0, and (G) sis-a1, sis- bsc3-1/FM7 stock at the non-permissive temperature of 29°C. Only 0–2, 2–3 and 5–7h embryos were analyzed in (C, E–G). SxlPe is most active in the 2–3h window. R2 antisense RNA shows a strong positive correlation with SxlPe expression (B and C), and is severely depressed in the background of reduced X chromosome counting genes of sis-a,b/FM7 (G). In all backgrounds, R1 as well as R2 sense RNA show an increase in the 5–7h window, which correlates with the shutdown of SxlPe. The AU level for the genotypes is slightly different; overall females have slightly reduced lncRNA levels and 29°C elevates transcription (* P-value <0.05, ** P-value <0.005, *** P-value <0.0005). Error bars represent mean +/− SEM.