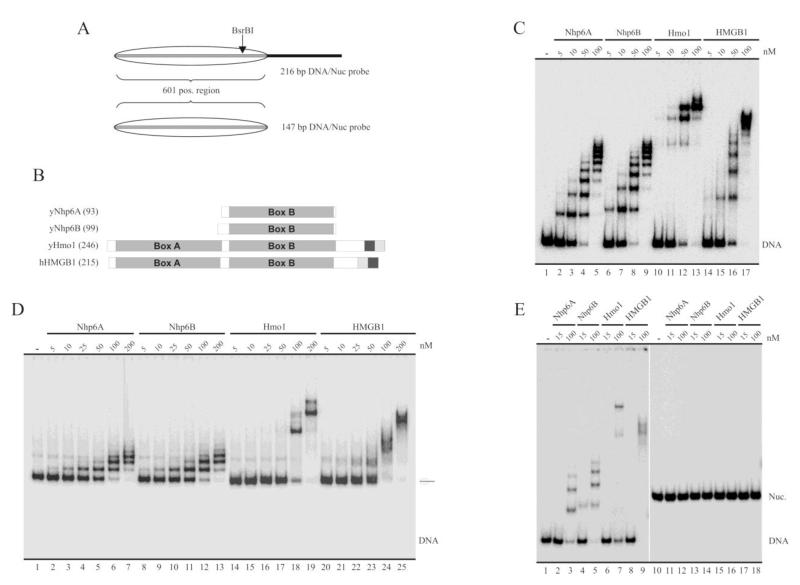
Fig. 1.
Binding properties of the yeast HMGB proteins under study. A, Schematic representation of the probes used in the analyses, represented here at the form of nucleosomal DNA. The transparent oval represents the position adopted by the histone octamer after reconstitution. 601 pos. region = nucleosome positioning sequence 601 (147 bp). B, Schematic representation of the proteins studied in the present work. The number of amino acids of each protein is indicated in parenthesis. The large gray squares termed Box A and Box B indicate the location of HMG-box domains. The dark gray and soft gray squares represent the acidic and basic stretches (respectively) present in the C-terminal region of these proteins. C-E, Electrophoretic analyses of binding patterns of the HMG proteins using non-denaturing polyacrylamide gels (5%, AA:Bis 40:1). The assays in (C) and (D) were performed using the 216 bp probe as naked DNA (C) or as a reconstituted mononucleosome (D). The assay in (E) used the 147 bp probe at the form of naked DNA (lanes 1-9) or as a reconstituted mononucleosome (lanes 10-18). The HMG protein used in each particular reaction is depicted on top of the figures, as well as the different concentrations tested.