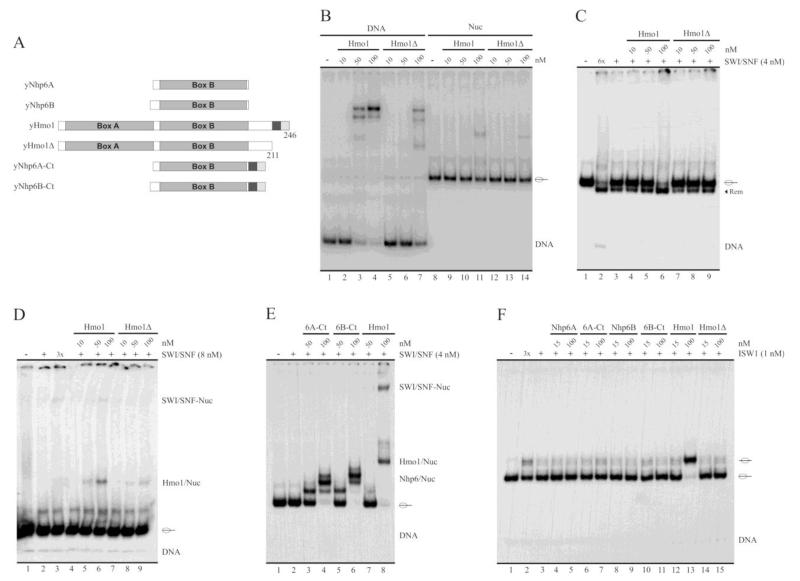
Fig. 5.
Hmo1’s stimulation of SWI/SNF remodeling activity and binding to the nucleosome is dependent on the C-terminal tail of this HMG protein. A, Schematic representation of WT Hmo1, its deletion mutant (Hmo1Δ), WT Nhp6A, WT Nhp6B and Nhp6 chimeric versions. Hmo1Δ corresponds to a C-terminal deletion mutant of Hmo1, lacking residues 212 to 246. Nhp6A-Ct and Nhp6B-Ct correspond to this 35 residues stretch fused to the C-terminal end of Nhp6A and B, respectively. The dark gray and soft gray squares represent the acidic and basic stretches (respectively) characteristic of Hmo1. B, Analysis of binding strength of Hmo1 and Hmo1Δ to the naked DNA and nucleosome probes, determined by electrophoresis in a non-denaturing polyacrylamide gel (5%, AA:Bis 40:1). C, Stimulation of SWI/SNF sliding activity by Hmo1 and Hmo1Δ. See legend of figure 2 for a general description of the assay. D, Analysis of stimulation of SWI/SNF binding to the nucleosome by Hmo1 and Hmo1Δ. E, Analysis of stimulation of SWI/SNF binding to the nucleosome by the chimeric forms of Nhp6A and Nhp6B, compared to Hmo1. See legend of figure 4 for a general description of the assays shown in (D) and (E). F, Stimulation of ISW1a sliding activity by WT and chimeric forms of Hmo1, Nhp6A and Nhp6B. See legend of figure 2 for a general description of the assay. The assays corresponding to figures B-D are representative of at least three independent assays. Migration of naked DNA is indicated at right of the pictures, where migration of the reconstituted mononucleosome is indicated schematically. The term “Rem” stands for remodeled (slid) nucleosome.