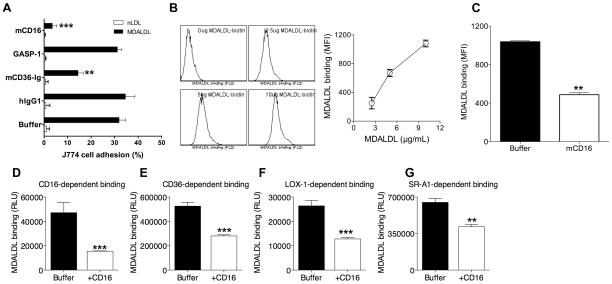
Figure 1. Mouse CD16 blocks MDALDL binding to macrophages.
A, CD16 inhibits J774 adhesion. MDALDL-coated ELISA plates were treated with indicated reagents (at 2 μg/ml) or buffer for 30 min. Calein-labeled J774 cells were added to the plates and incubated at 4°C for 30 min. After removing non-adherent cells by inverting the plates in PBS for 5 min, cell adhesion (%) was calculated by dividing fluorescence before and after washing the plate to remove non-adherent cells. B, Dose dependent MDALDL binding to RAW264 cells. RAW264 cells were incubated with MDALDLbiotin at indicated concentration for 1 h at 4°C and binding followed by streptavidin-PE. Cells incubated without MDALDLbiotin was used as a negative control. MDALDL binding was determined by FACS analysis. C, CD16 inhibits MDALDL binding to RAW264 macrophages. MDALDLbiotin (10 μg/ml) was preincubated without (buffer) or with sCD16 (5 μg/ml) for 2 h followed by its binding to RAW264 cells. D–G, Recombinant sCD16 blocks MDALDL binding to CD16. sCD16 or indicated soluble scavenger receptors were coated onto ELISA white plate. MDALDLbiotin was preincubated with 2.5 μg/ml sCD16 or buffer for 2 h and added to sCD16- (D) or sCD36 (E), sLOX-1 (F) or sSR-A- (G) coated plates. Binding was detected using streptavidin-alkaline phosphatase and Lumiphos 530 luminescence substrate as described in methods. In all these experiments values are means ± SD triplicate wells. Shown is a representative of three independent experiments. P, <0.01** or <0.001*** compared to buffer treated wells (no competitor).