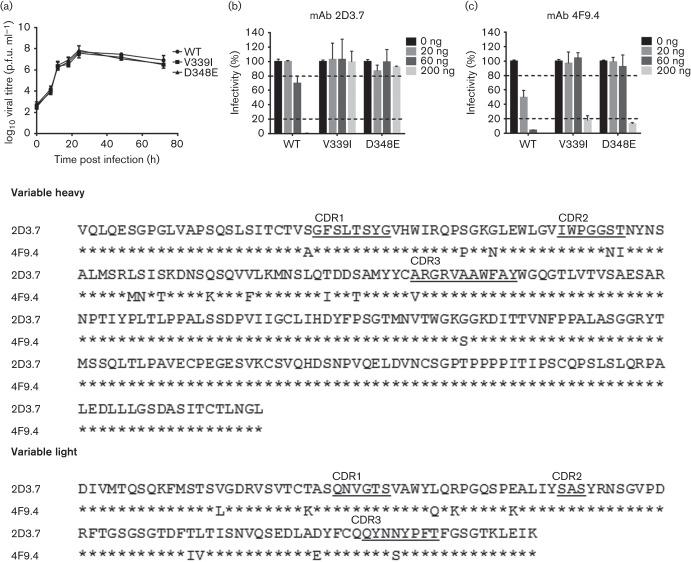
Fig. 7.
Characterization of 2D3.7 and 4F9.4 neutralization escape mutants. (a) RAW 264.7 cells were infected with the indicated viruses at m.o.i. of 2 on ice for 1 h. The inoculum was removed and cells were infected until indicated time points. Virus titres were determined by plaque assay. (b, c) Indicated recombinant viruses were subjected to in vitro neutralization by mAbs 2D3.7 (b) and 4F9.4 (c). Virus was incubated with the indicated concentrations of mAbs 2D3.7 and 4F9.4 for 30 min at 37 °C before performing plaque neutralization assay. The percentage infectivity was calculated by setting WT MNV without mAb as 100 %. The dashed lines indicate 20 and 80 % infectivity. Data are presented as means±sem of three independent experiments. (d) Sequence alignment of the heavy and light chain sequences for mAbs 2D3.7 and 4F9.4. The amino acid sequence of 2D3.7 is shown. Identical amino acids of 4F9.4 are indicated by asterisks. The complementary determining region (CDR) sequences are underlined.