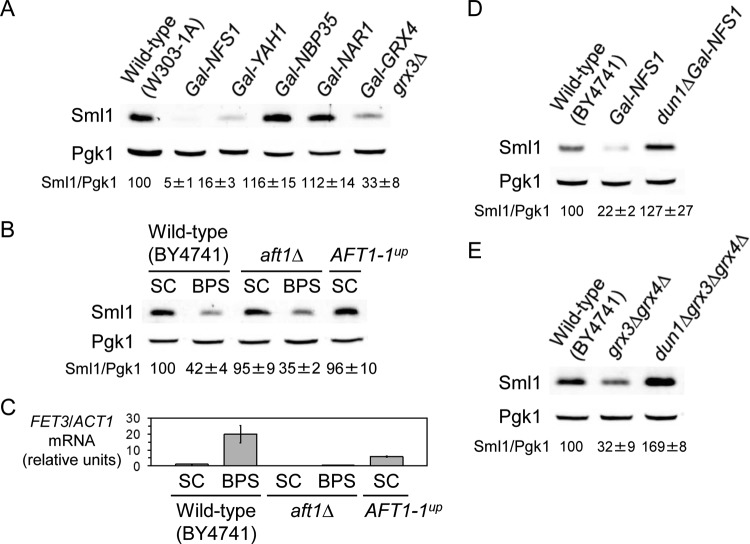
FIG 5.
Identification of genes involved in the regulation of Sml1 protein levels by low iron. (A) Sml1 protein abundance decreases in cells defective in members of the Fe deficiency-sensing pathway. Wild-type W303-1A, Gal-NFS1, Gal-YAH1, Gal-NBP35, Gal-NAR1, and grx3ΔGal-GRX4 yeast strains were grown at 30°C for 40 h in SC medium to repress the expression of the GAL-driven genes. As described in Materials and Methods, the cells were maintained in exponential phase during the whole incubation. (B) Sml1 protein levels in yeast strains with altered Aft1 activity. Wild-type BY4741, aft1Δ, and aft1Δ transformed with plasmid pAFT1-1up were grown at 30°C for 6 h in SC medium or SC medium with 100 μM BPS. (C) FET3 mRNA levels in yeast strains with altered Aft1 activity. Yeast cells were grown as for panel B, total RNA was extracted, and FET3 mRNA levels were determined by quantitative RT-PCR as described in Materials and Methods. FET3 mRNA values were normalized with ACT1 mRNA. (D) Dun1 kinase is required for the Sml1 protein drop observed in Gal-NFS1 cells. Wild-type BY4741, Gal-NFS1 (AXY1988), and dun1ΔGal-NFS1 (AXY2059) cells were grown as described in the legend to panel A. (E) Dun1 kinase is required for the Sml1 protein drop observed in grx3Δ grx4Δ cells. Wild-type BY4741, grx3Δ grx4Δ, and dun1Δ grx3Δ grx4Δ (AXY1926) cells were grown at 30°C in SC medium to exponential growth phase. In panels A, B, D, and E, Sml1/Pgk1 protein levels were determined as described in the legend to Fig. 1 and are relative to the values obtained for the wild-type strain grown in SC medium. In all cases, a representative experiment and the average and standard deviation from at least three independent biological replicates are represented.