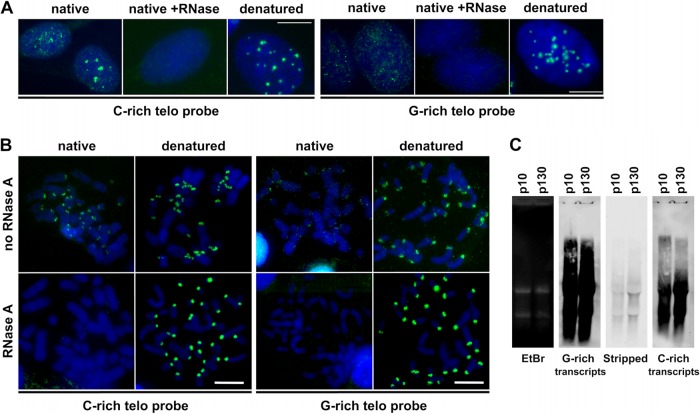
FIG 9.
Both G-rich and C-rich telomere strands are expressed in S. granarius cells. (A) RNA FISH experiments with strand-specific probes on S. granarius interphase nuclei. The C-rich probe detected either the UUAGGG or TTAGGG sequence, and the G-rich probe detected either CCCUAA or CCCTAA, depending on whether the hybridization was done on native or denatured nuclei, respectively. Signals were visible with both probes under native conditions without RNase treatment, albeit at lower intensity when the probe was G-rich. The signal was completely lost when native preparations were treated with RNase. Both probes detected telomeres equally efficiently under denaturing conditions. Nuclei are stained with DAPI (blue). (B) Similar RNA FISH experiment on metaphase chromosome preparations obtained by cytospin centrifugation under native or denatured conditions and treated or not treated with RNase. Once again, the C-rich probe yielded stronger signals than the G-rich probe, only on native, RNase-untreated chromosomes. Bars, 10 μm. (C) Northern blot analysis of S. granarius telomeric RNA transcripts in cells at different passages (p). (Left) Visualization of total RNA in the gel by ethidium bromide (EtBr) staining before transfer onto a membrane. (Second panel) Hybridization with a C-rich telomere probe labeled with digoxigenin. (Third panel) Verification of the stripping efficiency. (Right) Hybridization with a G-rich probe labeled with digoxigenin.