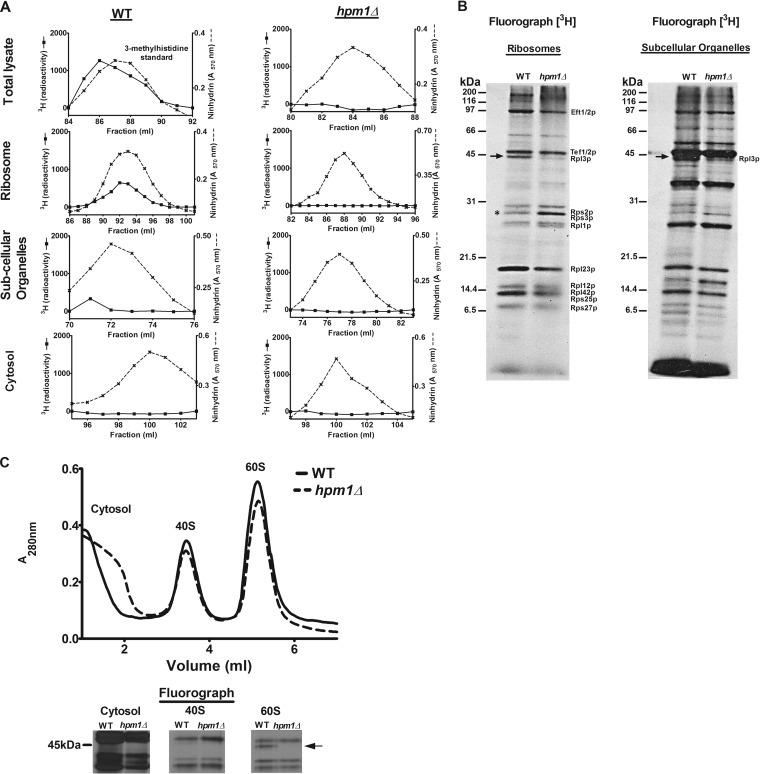
FIG 1.
Yeast protein 3-methylhistidine is found primarily on a ribosomal protein corresponding to Rpl3p and is dependent on HPM1. (A) Total lysate, ribosomes, subcellular organelles, and cytosol were prepared from WT (BY4742) and hpm1Δ mutant cells (BY4742) for amino acid analysis as described in Materials and Methods. The radioactivity shown (in counts per minute) has been normalized by background subtraction. Ninhydrin absorbance at 570 nm was measured to detect the 3-methylhistidine standard. (B) In vivo 3H-radiolabeled ribosomes and subcellular organelles from WT and hpm1Δ mutant cells (40 μg of protein) were pretreated with 2 U of Benzonase (Novagen 70746-4) for 30 min at 37°C and resolved by SDS-PAGE. 3H-methylated proteins were detected by fluorography for 13 weeks at −80°C as described in Materials and Methods. The arrow shows the methylated polypeptide corresponding to the molecular weight of Rpl3p, and the asterisk shows the positions of Rps2 and Rps3, whose levels of methylation are altered in the absence of HPM1. Radiolabeled bands corresponding to known methylated ribosomal proteins are identified on the right (14–22). (C) Seven OD units of WT and hpm1Δ mutant cells was radiolabeled with [3H]AdoMet, and the small 40S and large 60S ribosomal subunits were dissociated as described in Materials and Methods. One-hundred-microliter volumes of the cytosol, 40S, and 60S peak fractions were resolved by SDS-PAGE as described in Materials and Methods, except that a 12% Bis-Tris gel was prepared and run with MOPS running buffer. 3H-methylated proteins were detected by fluorography for 10 weeks at −80°C as described in Materials and Methods. At the bottom are three portions of lanes from the same gel.