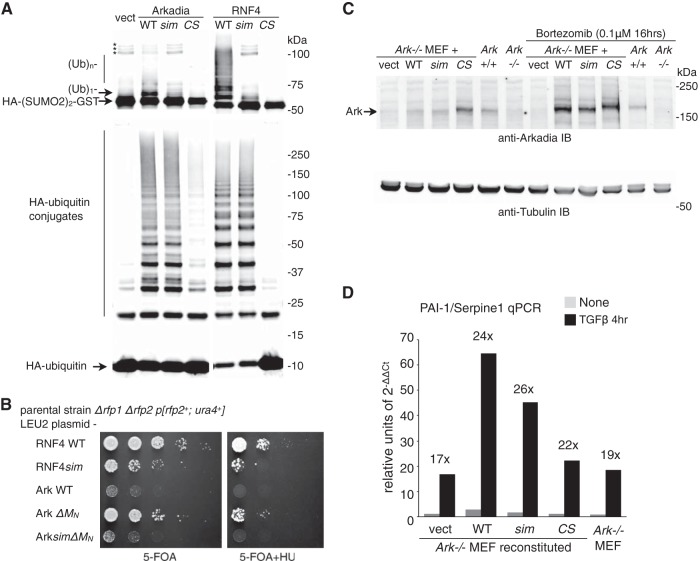
FIG 1.
Function of Arkadia's SUMO-binding domain is significant in vitro but insignificant in vivo. (A) SUMO-targeted ubiquitin ligase (STUbL) activity of Arkadia in an immunoprecipitation-coupled ubiquitin ligase assay. Flag-tagged Arkadia and its mutant forms (sim, sim13m [core hydrophobic residues, V298/V299/V300 in SIM1 and V380/V381/L383 in SIM3, changed to A]; CS, RING domain C966S mutant [change of C to S at position 966]) were immunoprecipitated in duplicates. The beads bound with the intact immune complex were used as the source for E3 in a ubiquitin ligase assay in vitro. The assay was conducted either with free ubiquitin (Ub) and an HA-tagged di-SUMO2–GST fusion protein as the substrates (top) for SUMO-targeted ubiquitin ligase (STUbL) activity or by using HA-tagged ubiquitin (bottom) for unspecified ligase activity. For both reactions, the products of the reaction mixture were analyzed with anti-HA antibody immunoblotting. *, nonspecific bands, likely aggregates of HA-(SUMO2)2-GST. (B) Results of plasmid shuffle, a yeast-based assay to examine the STUbL activity of RNF4, Arkadia, and their mutants (as indicated) in supporting the growth of the Δrfp1Δrfp2 double-mutant strain of fission yeast Schizosaccharomyces pombe. RNF4sim, sim23m mutant of RNF4, with core hydrophobic residues, I50/V51/L53 in SIM2 and V63/V63/L65 in SIM2, changed to A; 5-FOA, 5-fluoroorotic acid; HU, hydroxyurea. (C) Retrovirus-reconstituted expression of mouse Arkadia proteins in Ark−/− mouse embryo fibroblasts. The reconstituted expression of Arkadia proteins (as indicated), as well as the expression of endogenous Arkadia in wild-type (Ark+/+) MEFs, was determined through anti-Arkadia antibody immunoblotting. The amount of Arkadia proteins with an intact RING domain is increased upon proteasome inhibitor (bortezomib) treatment. The expression of α-tubulin in the lysates was shown as a loading control. (D) TGFβ signaling in wild-type and reconstituted Ark−/− MEFs. Expression of endogenous PAI-1/Serpine1 before and after a 4-h TGFβ stimulation was examined with RT-qPCR. Data are shown in arbitrary units based on the expression level of PAI-1/Serpine1 relative to that of GAPDH (2−ΔΔCt, cycle threshold). Cell lines used in the experiment are as indicated.