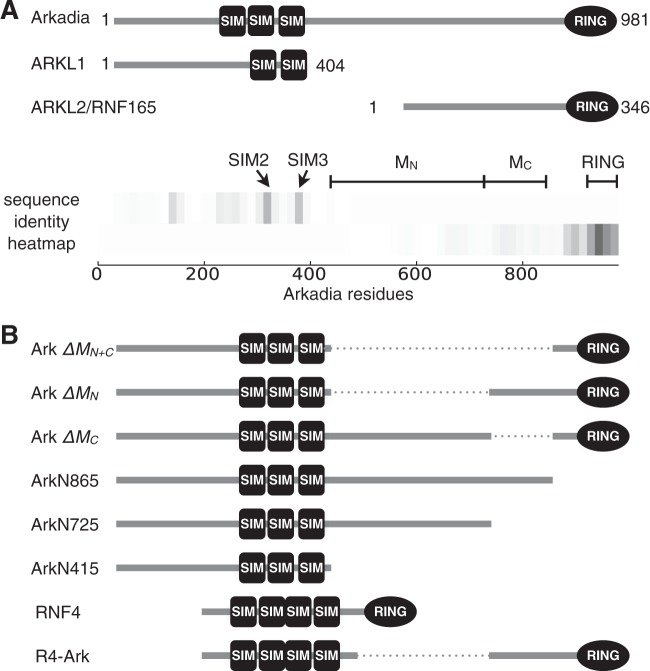
FIG 3.
Arkadia and the homologous Arkadia-like proteins (ARKL1 and ARKL2) are distinct in the middle region (M) of Arkadia between the SUMO-interacting motifs and the RING domain. (A) Domain structure of Arkadia, ARKL1, and ARKL2, with conserved SIMs and RING domain. A heat map (bottom) illustrates the conserved regions between the N-terminal half of Arkadia and ARKL1 and between the C-terminal half of Arkadia and ARKL2. The heat map was drawn using an array of identity scores assigned to each residue in Arkadia, based on the degree of local sequence identity it shares with ARKL1 or ARKL2 in a ClustalW alignment (actual alignment not shown); the gray scale (darkness) in the heat map corresponds to the degree of local sequence identity. The highly conserved SIM2, SIM3, and RING domain are as indicated. MN and MC denote two parts of the Arkadia M domain subject to mutational analyses in this study, described in the legend to panel B. (B) Diagrams of Arkadia mutants used in this study. In particular, ArkΔMN+C, ArkΔMN, and ArkΔMC refer to the deletion of residues 415 to 865, 415 to 725, or 725 to 865, respectively, in Arkadia; ArkN865 and ArkN725 are truncation mutants lacking the C terminus; R4-Ark is a chimeric fusion protein with the N-terminal 125 residues of RNF4 and the C-terminal 256 residues of Arkadia.