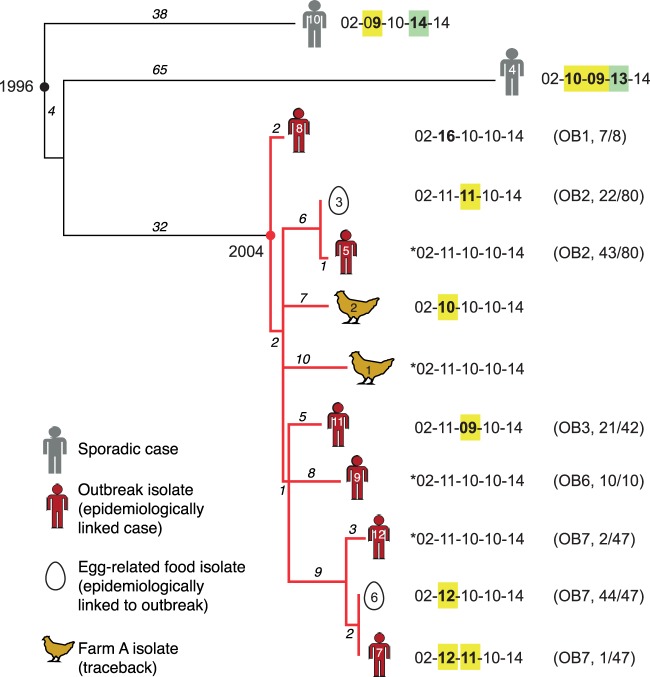
FIG 2.
Whole-genome phylogeny of 12 S. Typhimurium 135@ isolates from Tasmania. Numbers on branches indicate the number of SNPs (base changes) on each branch; divergence dates were estimated previously from SNP data using BEAST (30). Red branches indicate the outbreak clone. Each isolate is labeled with respect to its STm number (within the icon indicating specimen type), its MLVA profile, the outbreak from which it derives (OB [as described for Fig. 1]), and the proportion of isolates from that outbreak that share this profile. *, most-common MLVA profile, inferred to be ancestral to the outbreak clone. Copy number changes relative to this profile are highlighted in yellow and green. (Adapted from BMC Genomics [30] with additional annotation.)