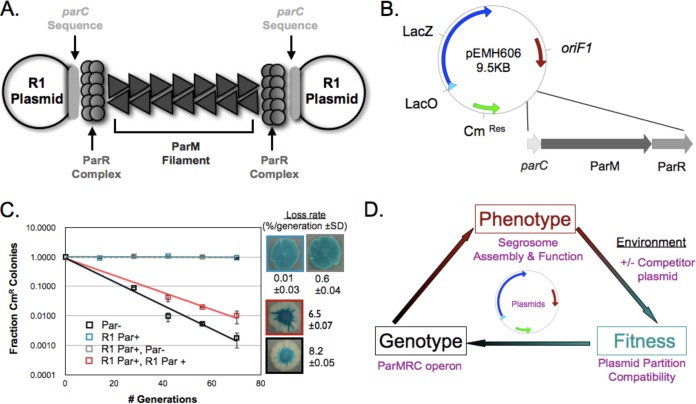
FIG 1.
Establishing a plasmid system, based on plasmid partitioning, to study molecular adaptation. (A) Schematic of the structure of a functional example of R1 partition machinery: the molecular machine encoded by the Par operon that separates two R1 plasmids. (B) Map of the low-copy-number mini-F1 plasmid, pEMH606, which served as the backbone of the resident plasmids, showing the presence of chloramphenicol resistance (CmRes) and β-galactosidase (LacZ) genes, the origin of replication, oriF1, and the location where the type II partitioning operons were inserted. (C) Qualitative and quantitative plasmid stability and incompatibility assays to monitor (i) the ability of the R1 Par operon (blue) to stabilize pEMH606 (black) and (ii) the destabilization of a resident plasmid, made by cloning the R1 Par operon into pEMH606, by a high-copy-number challenge plasmid that also carries the R1 Par operon (red). Error bars represent the 95% confidence intervals derived from two biological replicates, assayed in triplicate, and loss rates are provided with standard deviations (SD). (D) The relationship between genotype, phenotype, and fitness of the plasmid segregation system.