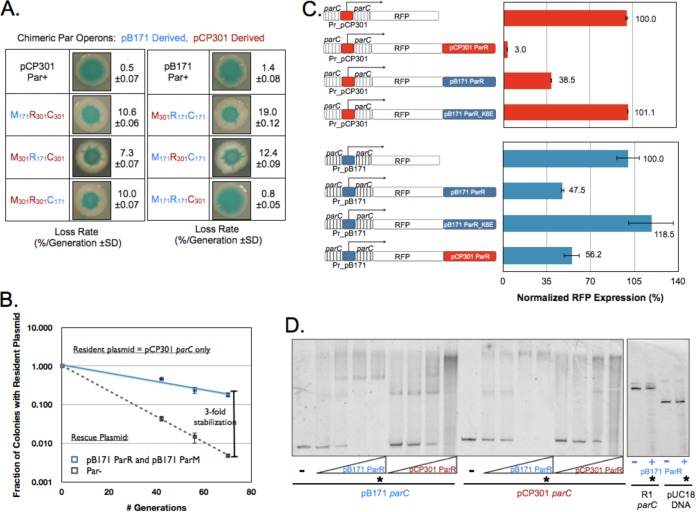
FIG 3.
Cross talk between the pB171 and pCP301 Par operons occurs at the ParR and parC interface. (A) A chimera with parCpCP301, ParRpB171, and ParMpB171 is stable. Data represent the results of qualitative and quantitative plasmid loss assays for resident plasmids carrying each of the 6 indicated chimeric operons as well as pB171 and pCP301 Par operons. (B) ParRpB171 and ParMpB171 stabilize resident plasmids containing only parCpCP301. Data represent the results of quantitative plasmid loss assays for a resident plasmid carrying only parCpCP301 in the presence of a rescue plasmid expressing either green fluorescent protein (GFP) (Par−) or ParRMpB171 from a strong constitutive promoter. Error bars represent the 95% confidence intervals derived from two biological replicates, assayed in triplicate. (C) ParRpB171 and ParRpCP301 each cross repress the other's parC. Genetic structure of eight RFP reporters, with RFP expressed from either the pCP301 parC promoter (Pr_pCP301; red bars) or the pB171 parC promoter (Pr_pB171; blue bars). Data represent the mean fluorescence of each strain normalized to that of the strain containing the appropriate, nonrepressed RFP reporter. Error bars represent the 95% confidence intervals derived from six independent transformants of each reporter plasmid. (D) pCP301 and pB171 each bind the other's parC in vitro. Data represent the results of a gel shift assay to assess the ability of purified His-tagged ParRpB171 and His-tagged ParRpCP301 protein to bind to both parCpB171 and parCpCP301 DNA. Lanes marked with a star contain equal amounts of purified ParRpB171 (8 μM) and a 2 nM concentration of the indicated DNA molecule. pUC18-derived DNA and parCR1 act as nonspecific control DNA sequences.