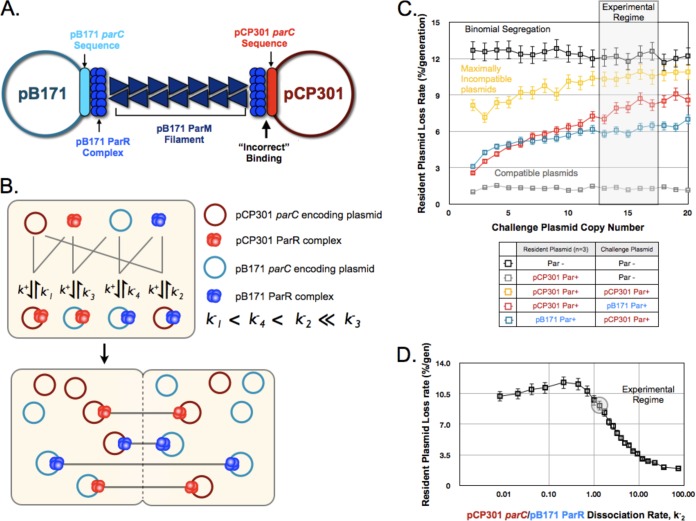
FIG 4.
A mixed-filament model can explain plasmid partition incompatibility. (A) Schematic of the proposed mixed filaments formed in cells containing both ParpCP301 and ParpB171 plasmids, illustrating the “incorrect” binding of ParRpB171 to parCpCP301. (B) A two-step kinetic model for pB171 and pCP301 incompatibility based on the relative in vivo binding affinities of each ParR protein complex to both parC sequences. See the supplemental material for details. (C) Output from the simulations of plasmid partition incompatibility for the indicated combinations of plasmids, indicating the dependence of the resident plasmid (copy number = 3) loss rate on the copy number of the challenge plasmid. The region corresponding to the experimentally determined mean copy number of the oriMB1 challenge plasmid is shaded. Error bars represent 95% confidence intervals. (D) The dependence of the kinetic model for pCP301 and pB171 incompatibility on k−2, the dissociation rate of ParRpB171 binding to parCpCP301. The resident plasmid and challenge plasmid copy numbers were set at 3 and 15, respectively, and the experimentally approximated value for k−2 is highlighted. Error bars represent the 95% confidence interval.