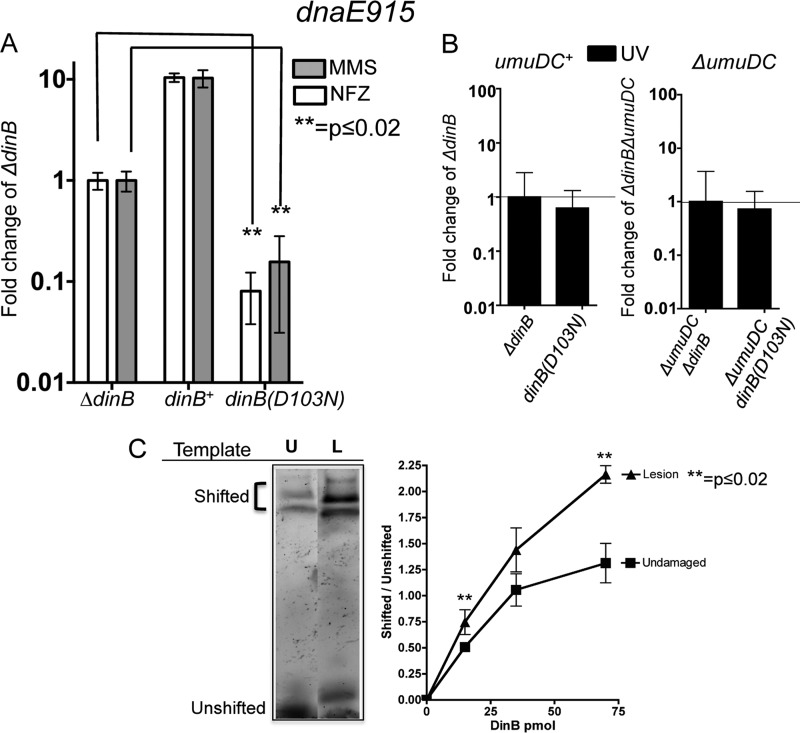
FIG 1.
DNA damage treatment results in survival loss only in strains with both chromosomally encoded antimutator dnaE915 and catalytically inactive dinB(D103N) alleles. (A) Treatment with either methyl methanesulfonate (MMS; 7.5 mM) or nitrofurazone (NFZ; 7.5 μM) of dnaE915 strains with the chromosomal catalytically inactive dinB allele [dinB(D103N)] results in hypersensivity to these DNA damage reagents. Fold changes were calculated with respect to the survival of a dnaE915 strain with a chromosomal deletion of dinB (ΔdinB) in NFZ (1.4 × 108 ± 1.5 × 107 CFU/ml) or in MMS (7.5 × 107 ± 2 × 106 CFU/ml). (B) Strains with chromosomal alleles dnaE915 and ΔdinB and with dnaE915 and dinB(D103N) equally survive UV treatment (∼55 J/m2) independent of the umuDC operon encoding DNA Pol V. Fold changes in this case were calculated based on UV treatment of dnaE915 ΔdinB (3 × 108 ± 3 × 107 CFU/ml) or ΔumuDC dnaE915 ΔdinB (3 × 107 ± 5 × 106 CFU/ml) strains. Error bars represent the standard deviations of the means from analyses of at least 3 independent isolates. (C) DinB prefers lesion-containing DNA. An electrophoretic mobility shift assay (EMSA) reveals that DinB preferentially binds to 3-deaza-3-methyl-adenine lesion-containing DNA (L) relative to undamaged DNA (U). Seventy-picomole DinB and 0.025-pmol DNA are shown. The lanes shown are from the same gel and have been electronically placed next to each other to directly compare the differences. The ratio of shifted (bound) to unshifted (unbound) DNA upon addition of the indicated concentrations of DinB is shown. Error bars represent the standard deviations of the means from analyses of at least 3 replicates. The band directly above the unshifted band in the L lane was not included in the quantification because we cannot consistently detect it. Band intensity was quantified using ImageJ (NIH).