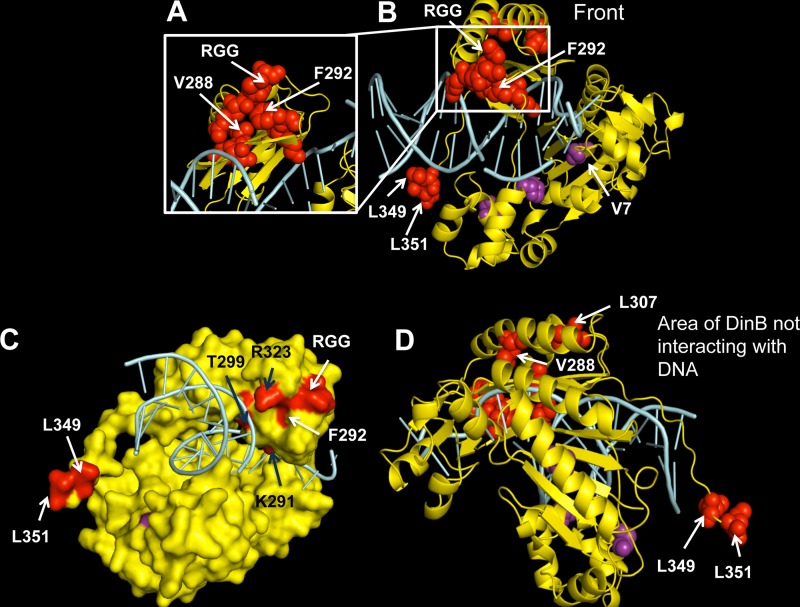
FIG 4.
A diverse array of intragenic dinB mutations suppresses survival loss. Shown is a mapping of the suppressor mutations on the in silico model of the DinB tertiary structure. Amino acid changes resulting from base pair substitutions and a three-amino-acid (arginine, glycine, glycine [RGG]) duplication identified in our assay are depicted as red or purple spheres in the DinB ribbon rendering. DinB protein is colored yellow, and DNA is colored cyan. Mutated residues mentioned in this paper (valine [V] 7 or 288, phenylalanine [F] 292, leucine [L] 307, 349, or 351; the RGG duplication) are labeled on the structure. (A) A magnified view of the area of DinB containing most amino acid mutations (including V288G and F292 to tyrosine [Y]). (B) Front view, defined here as the face of the DNA polymerase in which the DNA is also visible. (C) Surface rendering of DinB with mutations colored red or purple. The F292 amino acid is predicted to be on the surface of the protein. Black arrows indicate residues that might be interacting with DNA. (D) Depiction of the area of DinB that does not interact with DNA, the opposite face of that shown in panel B, in which we found no mutations. Images were rendered with PyMOL (DeLano Scientific, San Carlos, CA, USA).