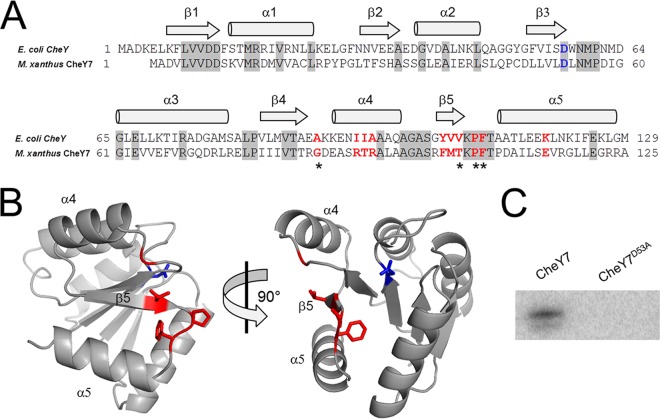
FIG 3.
CheY7 is a phosphorylatable SD-RR. (A) Amino acid alignment of E. coli CheY and M. xanthus CheY7. Residues important for interaction between E. coli CheY7 and FliM/CheZ substrates are indicated in red. The conserved aspartate residue that is the site of phosphorylation in REC domains is highlighted in blue. Gray shading indicates identity between E. coli and M. xanthus homologs. Regions of the proteins predicted to encode alpha helices or beta sheets are indicated above the alignment. (B) Depiction of two views of M. xanthus CheY7 threaded onto V. cholerae CheY3. Residues affecting CheY7-Cpc7 interactions as determined in the bacterial two-hybrid assay (Table 2) are shown in red. The putative site of phosphorylation is shown in blue. (C) In vitro phosphorylation of CheY7 and CheY7D53A using acetyl phosphate (see Materials and Methods) is shown. The D53A mutation eliminates phosphorylation of CheY7.