Abstract
Structural characterization of Streptococcus pneumoniae capsular polysaccharides (CPS) is a prerequisite for unraveling both antigenic and genetic relationships that exist between different serotypes. In the current study, comparative structural studies of S. pneumoniae CPS serogroup 10 (CPS10) were extended to include genetically related S. pneumoniae CPS34, CPS39, and CPS47F. High-resolution heteronuclear nuclear magnetic resonance (NMR) spectroscopy confirmed the published structure of CPS34 and, in conjunction with glycosyl composition analyses, revealed the following repeat unit structures of the other serotypes, which have not been previously characterized:

Common and unique structural features of these polysaccharides, including different positions of O-acetylation, were unambiguously associated with specific genes in each corresponding cps locus. The only exception involved the gene designated wcrC, which is associated with the α1-2 transfer of Gal pyranoside (Galp) to ribitol-5-phosphate in the synthesis of CPS10A, CPS47F, and CPS34 but with α1-1 transfer of Gal to ribitol-5-phosphate in the synthesis of CPS39. The corresponding gene in the cps39 locus, although related to wcrC, more closely resembled a previously identified gene (i.e., wefM) of Streptococcus oralis that is associated with α1-1 transfer of Galp to ribitol-5-phosphate. These and other recent findings identify linkages from α-Galp to ribitol-5-phosphate and from this residue to adjacent Gal furanoside (Galf) as important sites of CPS structural and genetic diversity.
INTRODUCTION
Streptococcus pneumoniae capsular polysaccharides (CPS) are of interest both as virulence factors and as protective immunogens for serotype-specific prevention of invasive disease. Currently recommended CPS-based vaccines include the 23-valent pneumococcal polysaccharide vaccine for adults and the 13-valent pneumococcal conjugate vaccine for children. Monitoring the efficacy of these vaccines requires ongoing surveillance of invasive pneumococcal serotypes (1). The traditional method for serotyping these bacteria is the Neufeld-Quellung test, which detects apparent swelling of bacterial capsules in the presence of factor antisera (2). Such determinations, although reliable, require training, experience, and specialized immunological reagents and, thus, are not amenable to widespread use. Molecular methods for serotyping these bacteria have also been described and are being developed that target genes for CPS biosynthesis in large operons (cps loci) located between dexB and aliA (3, 4). Sequencing of this region in reference strains of 88 S. pneumoniae serotypes revealed 1,502 genes, each defined by a different protein homology group (5). These include 430 genes for putative glycosyltransferases, 69 for sugar/polyalcohol phosphate transferases, 78 for acetyltransferases, 90 for different polymerases (Wzy) and 88 for different flippases (Wzx) (6). The proteins encoded by some of these genes have been shown biochemically to have the predicted function (4), and a number of others have been tentatively associated with specific steps in polysaccharide biosynthesis based on comparisons of CPS structure with cps gene content (6). The strength of the latter approach, however, is limited by the possibility of errors in existing polysaccharide structures, as well as by the absence of structures for many CPS serotypes.
S. pneumoniae serotypes were separated into eight genetic groups by cluster analysis of their cps loci (7). One of these groups (i.e., genetic cluster 4) included 23 serotypes, namely, the 15 serotypes that comprise CPS serogroups 10, 33, 35, and 47 and individual serotypes 13, 20, 29, 34, 36, 39, 42, and 43. Our interest in this group arose from the finding that certain so-called receptor polysaccharides (RPS) of Streptococcus oralis, an oral commensal, are closely related to S. pneumoniae CPS10F and other members of CPS serogroup 10 (8). Comparative structural studies of these RPS and CPS serotypes (9, 10) have now been extended to genetically related S. pneumoniae CPS34, CPS39, and CPS47F. Previous structural studies of these polysaccharides are limited to the chemical characterization of CPS34 (11–14). Confirmation of the CPS34 structure by nuclear magnetic resonance (NMR) and the determination of the CPS39 and CPS47F structures in the present study have allowed unambiguous association of the structure-determining genes in the cps loci of these serotypes with specific steps in polysaccharide biosynthesis.
MATERIALS AND METHODS
Glycosyl composition analysis.
All procedures associated with glycosyl composition analysis were performed at The University of Georgia Complex Carbohydrate Research Center. Initially, samples (400 μg) of CPS39 and CPS47F (Statens Serum Institute, Copenhagen, Denmark) were incubated with 48% aqueous hydrofluoric acid (HF) at 4°C for 48 h to cleave phosphodiester bonds, which were anticipated based on the presence of an encoded sugar/polyalcohol phosphate transferase (WhaI) in these serotypes (6). Samples were dried, and methyl glycosides prepared by methanolysis in 1 M HCl in methanol at 80°C (17 h), followed by re-N-acetylation with pyridine and acetic anhydride in methanol (for detection of amino sugars). Samples were then per-O-trimethylsilylated (TMS) by treatment with Tri-Sil (Pierce) at 80°C (0.5 h). Gas chromatography-mass spectrometry (GC-MS) analysis of TMS methyl glycosides was performed on an Agilent 7890A GC interfaced to a 5975C MSD (mass selective detector), using an Agilent DB-1 fused-silica capillary column (30 m by 0.25-mm inner diameter). Peaks were identified by comparison with monosaccharide standards, which included ribitol and arabitol.
NMR spectroscopy.
NMR studies were performed as previously described (10, 15) with native CPS39 and CPS47F and with de-O-acetylated samples of these polysaccharides that were prepared by overnight incubation of native CPS in mild base (0.1 M NH4OH) at 4°C. Native CPS34 was also examined. Prior to NMR, polysaccharides (3 to 10 mg) were lyophilized twice from 99.8% D2O and taken up in 99.996% D2O. Spectra were recorded at 20°C or 25°C in a Bruker DRX 500 spectrometer with a cryoprobe. A DRX700 spectrometer was used for 1H-detected 31P spectra. All proton and carbon chemical shifts were referenced relative to internal acetone using δ 1H = 2.225 ppm and δ 13C = 31.07 ppm.
Multiplicity-edited heteronuclear single quantum coherence (HSQC) was used to distinguish methylene from methine groups, which was useful for the identification of C-6 groups of hexoses, as well as C-1 and C-5 groups of pentitols in polysaccharides. The common homonuclear two-dimensional NMR methods of double-quantum-filtered coherence spectroscopy (DQF-COSY), total coherence spectroscopy (TOCSY), and nuclear Overhauser effect spectroscopy (NOESY) were augmented by triple-quantum-filtered coherence spectroscopy (TQF-COSY), which was useful for correlating methylene protons with adjacent positions. The utility of the hybrid method HSQC-TOCSY in the crowded carbohydrate spectra was enhanced by high digital resolution in the indirect dimension (13C). With an increased number of data points (2,048) and by folding of these single-quantum spectra, the useful resolution was improved to approximately 3 Hz, approaching the natural 13C line width. A second hybrid pulse sequence, HSQC-NOESY, also recorded at high digital resolution in 13C, was especially valuable for correlating C-5 of β-Gal pyranoside (β-Galp) with H-1 and H-4. Heteronuclear multiple bond correlation (HMBC) spectra were used for identifying linkage positions and assignments of residues. A variation of this sequence in which the 13C carrier was placed in the region of the carbonyl 13C (175 ppm) was used for reliable correlation of acetyl and amide methyl groups with proton resonances in the sugar ring. All NMR data were processed by using NMRpipe and NMRDraw (NMRScience), with analysis using NMRview (One Moon Scientific) and Sparky software.
RESULTS
Structure of CPS39.
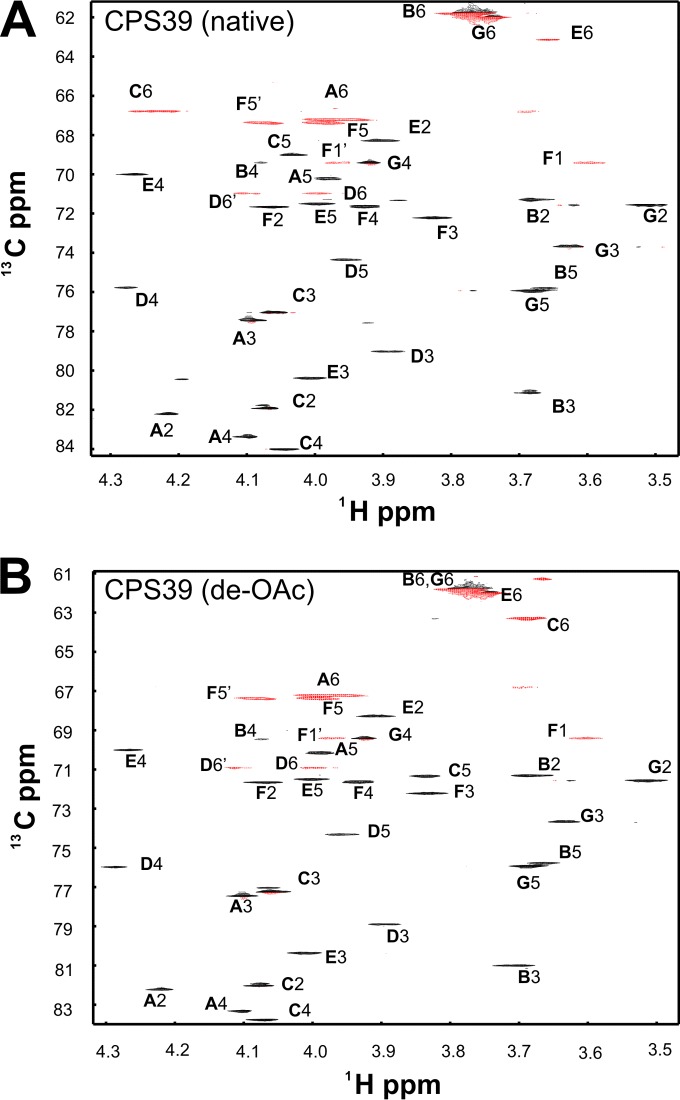
Glycosyl composition analysis of HF-treated CPS39 revealed the molar composition to be 76.5% Gal, 13.2% GalNAc, and 9.7% ribitol. While we did not include analysis of the absolute configuration of the residues, it is clear from the homologies between the genes for the glycosyl transferases of CPS39 and the serogroup 10 and 34 CPSs that the residues are all the common d configuration. The presence of six sugar residues in the repeating subunit of both native CPS39 and de-O-acetylated CPS39 was suggested by six resonances in the anomeric region (from 100 to 110 ppm in 13C and 4.4 to 5.2 ppm in 1H) of multiplicity-edited HSQC spectra (see Fig. S1 in the supplemental material). Other resonances in this region included one that was indicative of an acetamido sugar at 53 ppm 13C and 4.16 ppm 1H and two 1H resonances of the same intensity that were characteristic of acetyl methyl groups. One acetyl methyl resonance (at 2.048 ppm in 1H and 23.04 ppm in 13C) is from the amide and the other (at 2.147 ppm 1H and 21.22 ppm 13C) is from an O-acetyl substituent of a sugar residue, as indicated by its absence in the spectrum of de-O-acetylated CPS39 (see the text in the supplemental material). The position of the O-acetyl group in CPS39 is established as described below from complete assignments of the NMR spectra of native and de-O-acetylated polysaccharides.
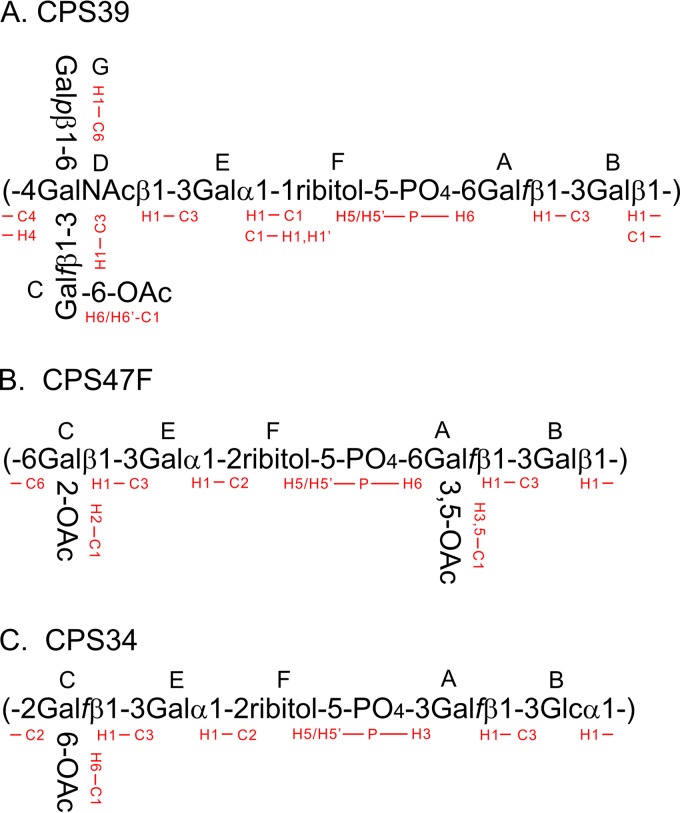
Our assignment of native CPS39 established the identities and anomeric configurations of the component sugar residues, as well as the positions of linkages (Fig. 1). We give a brief overview of the assignment strategy here; a more detailed explanation is given in the text in the supplemental material. We began by assigning an underlined capital letter (as defined in Fig. 2) to the six anomeric sugar residues, similar to the boldface capital letters used in previous NMR studies of CPS serogroup 10 (9, 10). Both the genes in the cps39 locus and our glycosyl composition analysis indicate a ribitol residue that we will describe as residue F in our analysis of the NMR data. In addition to the major NMR signals we discuss, there are several minor signals at levels of 10 to 15% in the anomeric region, indicating a modest level of contamination of the commercial CPS sample by unknown polysaccharides whose presence does not influence any of the conclusions drawn in this study. Residues A and C were identified as β-Gal furanoside (β-Galf) by the chemical shifts of C-1, C-2, C-3, and C-4, which appeared at positions that were relatively downfield in the 13C dimension (Fig. 1A, Table 1). The associated atoms likewise showed characteristic 1H homonuclear and 1H-13C coupling patterns typical of β-Galf. Residues B and G were identified as β-Galp by their homonuclear JHH coupling values that indicated axial H-1, H-2, and H-3 with equatorial H-4. Residue D was identified as a β-GalNAc pyranoside by the same pattern of JHH as β-Galp but with a 13C chemical shift for the carbon at position 2 of residue D (D2-C) (52.97 ppm) that is typical of a carbon replaced by a nitrogen atom. Residue E was recognized as α-Galp by a small value of JH1-H2 and a large value of JH2-H3, indicating an equatorial H-1 of an α-pyranoside; the small coupling constants of E4-H indicated an equatorial proton. All remaining resonances were assigned to residue F (ribitol).
FIG 1.
Central region of the multiplicity-edited 1H-13C HSQC spectra of native CPS39 (A) and de-O-acetylated CPS39 (B). Negative contours (red) indicate methylene groups.
FIG 2.
Structures and HMBC interresidue connectivities of CPS39 (A), CPS47F (B), and CPS34 (C).
TABLE 1.
Residue-by-residue comparison of HSQC 1H and 13C chemical shifts for S. pneumoniae CPS10A, CPS39, CPS47F, and CPS34 and S. oralis RPS4Gna
| Polysaccharide | Residue | Structure | Chemical shifts (ppm)b |
|||||
|---|---|---|---|---|---|---|---|---|
| H-1 C-1 | H-2 C-2 | H-3 C-3 | H-4 C-4 | H-5 C-5 | H-6 C-6 | |||
| CPS10A | A | 5-β-Galf | 5.225 110.17 | 4.215 82.73 | 4.189 77.30 | 4.210 82.67 | 4.378 75.77 | 3.784, 3.862c 62.82 |
| CPS39 | 6-β-Galf | 5.223 110.17 | 4.216 82.23 | 4.096 77.46 | 4.099 83.37 | 3.981 70.22 | 3.966 67.22 | |
| CPS39deOAcd | 6-β-Galf | 5.227 110.12 | 4.216 82.22 | 4.098 77.48 | 4.099 83.34 | 3.989 70.15 | 3.953, 3.983 67.22 | |
| RPS4Gn | 6-β-Galf | 5.218 110.05 | 4.208 82.26 | 4.101 77.46 | 4.108 83.36 | 3.990 70.10 | 3.970 67.18 | |
| CPS47Fd | 6-β-Galf | 5.301 110.04 | 4.359 79.61 | 4.88 79.75 | 4.498 81.14 | 5.385 71.84 | 4.051 64.40 | |
| CPS47FdeOAc | 6-β-Galf | 5.217 110.08 | 4.210 82.26 | 4.096 77.47 | 4.102 83.40 | 3.983 70.16 | 3.950, 3.981 67.19 | |
| CPS34 | 3-β-Galf | 5.302 109.15 | 4.354 81.02 | 4.473 81.58 | 4.301 83.94 | 3.947 71.29 | 3.699 63.80 | |
| CPS10A | B | 3-β-Galp | 4.760 104.41 | 3.692 71.73 | 3.692 81.8 | 4.085 69.84 | 3.656 76.06 | 3.762 62.24 |
| CPS39 | 3-β-Galp | 4.764 103.94 | 3.683 71.28 | 3.684 81.15 | 4.082 69.41 | 3.669 75.80 | 3.768, 3.788 61.81 | |
| CPS39deOAc | 3-β-Galp | 4.767 104.00 | 3.676 71.30 | 3.704 81.03 | 4.070 69.45 | 3.666 75.76 | 3.769 61.84 | |
| RPS4Gn | 3-β-Galp | 4.507 103.91 | 3.675 70.84 | 3.742 81.09 | 4.107 69.38 | 3.730 75.96 | 3.78 61.85 | |
| CPS47F | 3-β-Galp | 4.506 103.90 | 3.666 70.78 | 3.761 80.90 | 4.083 69.25 | 3.718 76.08 | 3.777 61.85 | |
| CPS47FdeOAc | 3-β-Galp | 4.504 103.87 | 3.668 70.86 | 3.737 81.10 | 4.101 69.37 | 3.724 75.95 | 3.769, 3.788 61.85 | |
| CPS34 | 3-α-Glcp | 5.093 98.56 | 3.724 72.03 | 3.859 79.60 | 3.484 68.64 | 3.807 73.19 | 3.79, 3.88 61.48 | |
| CPS10A | C | β-Galf | 5.073 110.52 | 4.078 82.53 | 4.055 77.92 | 4.068 84.68 | 3.841 72.02 | 3.684 63.93 |
| CPS39 | β-Galf | 5.062 110.06 | 4.069 81.93 | 4.064 77.07 | 4.048 84.01 | 4.036 69.01 | 4.220, 4.240 66.79 | |
| CPS39deOAc | β-Galf | 5.056 110.06 | 4.073 82.09 | 4.055 77.23 | 4.068 83.77 | 3.834 71.32 | 3.676 63.28 | |
| RPS4Gn | 6-β-Galf | 5.071 108.62 | 4.068 81.81 | 4.100 77.51 | 4.006 83.96 | 4.027 70.78 | 3.767, 4.077 71.91 | |
| CPS47F | 6-β-Galf | 5.386 107.89 | 5.061 84.78 | 4.239 76.56 | 4.151 84.78 | 4.049 70.64 | 3.780, 4.069 71.78 | |
| CPS47FdeOAc | 6-β-Galf | 5.231 109.94 | 4.207 82.24 | 4.077 77.65 | 4.069 83.90 | 4.022 70.71 | 3.771, 4.074 71.84 | |
| CPS34 | 2-β-Galf | 5.362 108.34 | 4.271 87.35 | 4.270 76.06 | 4.103 82.55 | 4.082 68.61 | 4.247 66.46 | |
| CPS10A | D | 3,4,6-β-GalNAc | 4.788 103.76 | 4.145 53.54 | 3.928 79.29 | 4.269 76.51 | 3.938 74.84 | 3.979, 4.099 71.22 |
| CPS39 | 3,4,6-β-GalNAc | 4.747 103.60 | 4.162 52.97 | 3.896 79.04 | 4.279 75.78 | 3.955 74.37 | 3.992, 4.097 70.98 | |
| CPS39deOAc | 3,4,6-β-GalNAc | 4.74 103.61 | 4.159 52.98 | 3.894 78.94 | 4.286 75.98 | 3.958 74.32 | 3.997, 4.096 70.93 | |
| RPS4Gn | 6-β-GalNAc | 4.635 103.93 | 3.944 53.36 | 3.752 71.56 | 3.945 68.59 | 3.829 74.54 | 3.775, 3.911 68.08 | |
| CPS10A | E | 3-α-Galp | 5.203 100.36 | 3.938 68.88 | 4.006 80.70 | 4.272 70.29 | 4.057 72.25 | 3.762 62.24 |
| CPS39 | 3-α-Galp | 4.962 99.93 | 3.898 68.30 | 4.005 80.38 | 4.263 70.01 | 3.997 71.49 | 3.744 62.01 | |
| CPS39deOAc | 3-α-Galp | 4.964 99.92 | 3.906 68.28 | 4.004 80.38 | 4.266 70.00 | 3.996 71.50 | 3.745 62.00 | |
| RPS4Gn | 3-α-Galp | 4.959 99.92 | 3.890 68.26 | 3.963 80.22 | 4.204 70.08 | 3.980 71.43 | 3.74 62.06 | |
| CPS47F | 3-α-Galp | 5.247 100.03 | 3.965 68.49 | 4.016 77.92 | 4.164 69.97 | 4.083 72.03 | 3.744 61.80 | |
| CPS47FdeOAc | 3-α-Galp | 5.25 99.95 | 3.962 68.50 | 3.977 78.17 | 4.168 69.98 | 4.082 72.02 | 3.733, 3.754 61.84 | |
| CPS34 | 3-α-Galp | 5.238 100.31 | 3.957 68.52 | 4.008 77.92 | 4.143 70.28 | 4.091 72.00 | 3.747 61.80 | |
| CPS10A | F | 2-Ribitol-5 | 3.856, 3.931 61.36 | 4.017 80.81 | 3.879 71.91 | 3.998 73.18 | 4.022, 4.145 68.33 | |
| CPS39 | 1-Ribitol-5 | 3.596, 3.965 69.42 | 4.066 71.66 | 3.828 72.22 | 3.928 71.64 | 3.989, 4.080 67.39 | ||
| CPS39deOAc | 1-Ribitol-5 | 3.601, 3.968 69.41 | 4.068 71.65 | 3.829 72.21 | 3.93 71.62 | 3.994, 4.076 67.37 | ||
| RPS4Gn | 1-Ribitol-5 | 3.599, 3.965 69.48 | 4.054 71.63 | 3.818 72.29 | 3.933 71.66 | 3.99, 4.085 67.40 | ||
| CPS47F | 2-Ribitol-5 | 3.845, 3.926 60.83 | 4.052 80.62 | 3.845 71.21 | 4.023 72.39 | 3.943, 4.057 67.83 | ||
| CPS47FdeOAc | 2-Ribitol-5 | 3.840, 3.928 60.80 | 4.052 80.57 | 3.859 71.23 | 4.031 72.38 | 3.977, 4.084 67.75 | ||
| CPS34 | 2-Ribitol-5 | 3.847, 3.922 60.77 | 4.047 80.80 | 3.869 71.24 | 4.044 72.35 | 4.008, 4.096 67.79 | ||
| CPS10A | G | β-Galp | 4.435 104.84 | 3.532 72.09 | 3.627 74.23 | 3.943 69.94 | 3.676 76.30 | 3.762 62.24 |
| CPS39 | β-Galp | 4.436 104.51 | 3.511 71.56 | 3.628 73.70 | 3.921 69.41 | 3.680 75.92 | 3.782 61.82 | |
| CPS39deOAc | β-Galp | 4.439 104.47 | 3.510 71.54 | 3.631 73.67 | 3.923 69.41 | 3.685 75.91 | 3.768, 3.784 61.84 | |
Based on homology between the cps loci of CPS serogroup 10 and CPS39, we anticipated the presence of a phosphodiester bond in the CPS39 repeat, and thus, we recorded 1H-31P HSQC spectra for this polysaccharide. These spectra showed cross peaks of different intensities at two different 31P chemical shifts. The more intense peaks were assigned to CPS39, which was the main constituent, while the peaks of lesser intensity arose from a low level of contaminating polysaccharide(s) containing phosphocholine. The HSQC spectrum of CPS39 contained two methylene resonances that were not assigned to hexoses. One of these resonances was assigned to F5 by the scalar coupling of F5,5′-H to 31P of the phosphodiester group. 31P-1H HSQC also showed coupling of the phosphate resonance to A6-H, thereby identifying a phosphodiester linkage between residue F (ribitol) and the adjacent residue A (Galf). The remaining atoms of the ribitol (F2-C, F3-C, and F4-C) were assigned with the assistance of TQF-COSY, which correlated F2-H with F1,1′-H and F4-H with F5,5′-H.
Once the assignment of all 13C-1H resonances of these polysaccharides was complete (Table 1), we verified that all resonances in the spectra (Fig. 1; see also Fig. S1 in the supplemental material) were accounted for. The complete structure of native CPS39 was determined from long-range C-H correlations between each anomeric proton and the linkage carbon atom or between the anomeric 13C and the linkage proton (Fig. 2A). Details of the correlations used to deduce the structure are discussed in the text in the supplemental material. The principal difference in chemical shifts of native and de-O-acetylated CPS39 is in C6 (compare Fig. 1A and B and values highlighted by boldface in Table 1); the 13C shift of the native form was 3.51 ppm downfield from that of the de-O-acetylated form. There was also a substantial shift of C6-H, accompanied by smaller changes at both C5-H and C5-C, suggesting that the position of the O-acetyl substituent is at position 6 of the Galf residue, C. This conclusion was supported by long-range 13C-1H correlation data from the carbonyl HMBC, as described in the text in the supplemental material.
Structure of CPS47F.
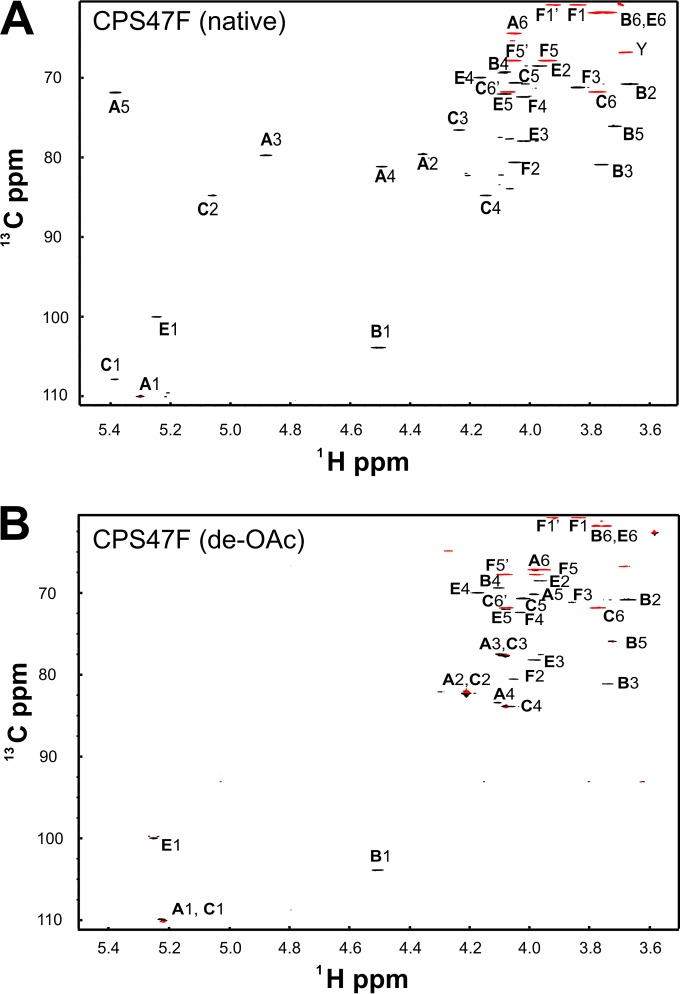
Glycosyl composition analysis of HF-treated CPS47F revealed the molar composition to be 85.1% Gal, with no significant GalNAc (0.8%), and 11.5% ribitol. The anomeric region of the HSQC spectrum of CPS47F (Fig. 3) contained four resonances, indicating four sugar residues in the CPS47F repeating unit. However, eight peaks were present in the corresponding region (between 5.4 and 4.4 ppm) of the 1H NMR spectrum (Fig. 3A). As described in the text in the supplemental material, four of the eight peaks were associated with extensive O-acetylation of CPS47F. The upfield region, characteristic of methyl groups, contained three peaks, the intensities of which were comparable to three times that of the anomeric peaks. These methyl groups could not be attributed to N-acetyl groups, as no significant GalNAc was detected by monosaccharide analysis of CPS47F and no peaks were present in the 50-ppm region of the 13C dimension of the HSQC spectrum that would indicate N-acetyl amino sugars. Thus, the three methyl groups detected were interpreted to indicate the presence of three O-acetyl groups per repeating subunit.
FIG 3.
Multiplicity-edited 1H-13C HSQC spectra of native CPS47F (A) and de-O-acetylated CPS47F (B). Negative contours (red) indicate methylene groups. The peaks marked Y are from minor contaminating polysaccharides.
Residue letters A, B, C, and E were assigned to the four anomeric sugars of CPS47F, following the notations used with CPS39 as indicated in Fig. 2. The arguments for assignment of resonances to residue E (α-Galp) in CPS47F closely followed those used for residue E in CPS39, as did those for assignment of resonances to residues A and C. Large differences between the chemical shifts of these polysaccharides were evident, however (Table 1). As explained in the text in the supplemental material, these differences resulted from O-acetylation of residue positions A3, A5, and C2 of CPS47F (compare Fig. 3A and B). This modification was established by carbonyl HMBC, which showed long-range correlation of the carbonyl carbons, both to methyl group protons and to ring protons A3-H, A5-H, and C2-H. The only difficulty encountered in the assignment of residue B (β-Galp) involved B1 through B4, which arose from the exact overlap of the chemical shift of B1-H with A4-H at 4.5 ppm. However, resonances from these residues were distinguished in HSQC-TOCSY spectra by the effect of O-acetylation on dispersion of the signals from residue A. B5-C was assigned by the HSQC-NOESY cross peak with B1-H. B6, which was assigned by HSQC-TOCSY, was only slightly resolved from E6.
After assignment of the four sugar residues, two methylene group resonances and F5-H remained that were assigned by 31P-1H HSQC. The assignment of F5 at (H,H′) 4.059, 3.943 ppm 1H, and 67.84 ppm 13C left F1 as the remaining methylene signal at 3.926, 3.845 1H, and 60.85 13C. F2, F3, and F4 were then assigned by HSQC-TOCSY. Given the complete assignment of the 1H and 13C spectra (Table 1), linkages between the residues (Fig. 2B) were determined by long-range C-H correlation spectra as described in the text in the supplemental material.
The chemical shift dispersion seen in the NMR spectrum of de-O-acetylated CPS47F was considerably less than that seen with native CPS47 (compare Fig. 3A and B). The anomeric 1H chemical shifts of residues A, C, and E, which were well separated with CPS47F (Fig. 3A), fell within in a 0.03-ppm range with de-O-acetylated CPS47F (Fig. 3B). Assignment of these residues by the use of HMBC and HSQC-TOCSY was complicated by the chemical shift overlap of both the 13C and 1H resonances of A2 and C2. Thus, HSQC-TOCSY cross peaks of the anomeric signals with the well resolved signals of A3-C and A4-C, as well as C3-C and C4-C, were used to assign all resonances (Table 1). Given the assignments of A1 to A4 and of C1 to C4, the assignments of A5, A6, C5, and C6 were readily apparent from HSQC-TOCSY. The resonances of residue E were quite similar between native and de-O-acetylated CPS47F (Table 1) and were assigned by the same correlations. Residue B (β-Galp) was assigned by HSQC-TOCSY cross peaks between B1-H and B2-C and between B3-C and B4-C, with data from COSY and HSQC-NOESY distinguishing B2 from B3. The small homonuclear splitting of B4-H identified this equatorial proton of galactose. The signal of B5-C failed to give a good HSQC-NOESY cross peak with B1-H, but it did give a peak with B4-H. HSQC-TOCSY identified the methylene resonance of B6-H, which overlaps that of E6, as was the case for the native CPS.
We knew from the structure of native CPS47F that E1 is linked through the 2 position of ribitol (residue F). Thus, the HMBC cross peak from E1-H to F2-C at 80.57 ppm served as a starting point for the assignment of residue F in de-O-acetylated CPS47F. HSQC-TOCSY was used to complete the assignment of residue F (ribitol). The linkages in de-O-acetylated CPS47F, as determined by HMBC and HSQC-NOESY, were all observed to be the same as in native CPS47F.
Structure of CPS34.
The available structure of CPS34 (16) is based on the elegant and extensive chemical studies of Baddiley and coworkers (11–13). To verify this structure, we recorded 1H and 13C NMR spectra and assigned all resonances to facilitate future identification of this polysaccharide.
Given the complete assignment of the 13C and 1H resonances of CPS34 (Table 1; see also the text and Fig. S2 in the supplemental material), the structure of the CPS was deduced from the HMBC spectrum, which showed correlations of A1-H with B3-C, B1-H with C2-C, C1-H with E3-C, and E1-H with F2-C, and from the 31P-1H HSQC spectrum, which showed that phosphate links F5 and A3. The structure of CPS34 determined here is essentially identical to the one determined previously (11) by chemical methods. The only minor difference is our finding of stoichiometric O-acetylation of Galf (residue C) at C-6 (see the text and Fig. S2 in the supplemental material) rather than ∼50% O-acetylation of the same residue at C-5 or C-6 (12).
DISCUSSION
The CPS structures determined here (Fig. 2), including the positions of O-acetyl groups, depend primarily on scalar (J) coupling correlations between spin 1/2 nuclei (1H, 13C, and 31P) rather than on either NMR chemical shift analogies or NOESY. This is important because, in carbohydrates, 1H-13C scalar coupling correlations follow the bonds that determine chemical structure, whereas chemical shift analogies are influenced by the complex stereochemistry and unpredictable geometry of carbohydrates and, thus, are not reliable predictors of structure. Likewise, NOESY, which indicates proximity between protons, provides a reliable indicator of stereochemistry for pyranosides of known sugar puckering but does not always accurately indicate the position of a linkage. All chemical shifts and NOE of the polysaccharides examined do, however, show the effects of glycosylation and O-acetylation that are expected from their proposed structures. Moreover, O-acetylation of these structures at the reported positions is close to stoichiometric based on NMR spectra recorded at room temperature and neutral pH, conditions that do not cause migration or hydrolysis of O-acetyl groups.
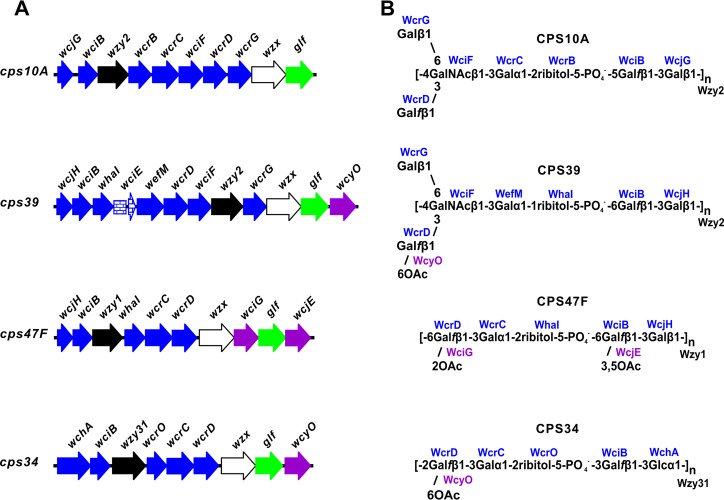
The structures of S. pneumoniae CPS39, CPS47F, and CPS34 are compatible with the well-established properties of several genes in the cps loci of these serotypes. These include wcjG, wcjH, and wchA for different initial transferases that link Galp or Glc to carrier lipid and wciB for the subsequent β1-3 transfer of Galf to either Galp or Glc (Fig. 4A). The transfer of Galf appears to create acceptors for four putative ribitol-5-phosphate transferases, each with a different linkage specificity (Table 2). Thus, wciO is associated with 5-P-2 linkages in both CPS6A and CPS6B (6) and CPS33B and CPS33D (17), wcrO with 5-P-3 linkages in CPS33C (17) and CPS34 (Fig. 4), wcrB with 5-P-5 linkages in all members of CPS serogroup 10 (9, 10, 18), and whaI with 5-P-6 linkages in CPS39 and CPS47F (Fig. 4B), as well as in CPS47A (19). The occurrence of these genes (i.e., wciO, wcrO, wcrB, and whaI) is limited to the 16 S. pneumoniae serotypes listed in Table 2. Thus, their association with CPS structure is nearly complete.
FIG 4.
Genetic basis of S. pneumoniae serotype 10A, 39, 47F, and 34 CPS structures. (A) Structure-determining regions of cps loci (GenBank accession numbers CR931649, CR931711, CR931721, and CR931703, respectively), showing genes for glycosyl or ribitol-phosphate transferases (blue), polymerases (black), flippases (white), O-acetyltransferases (purple), and galactofurannose mutase (green); wciE (blue bricks) occurs as a pseudogene in the cps39 locus. (B) Association of encoded transferases and polymerases (Wzy homology group 1, 2, or 31) with each corresponding CPS structure. Gene names are the same as in previous studies (5, 6) except for wcrC in the cps39 locus, which has been renamed wefM.
TABLE 2.
Specificity and serotype distribution of selected S. pneumoniae ribitol-5-phosphate transferases or glycosyltransferases inferred from known CPS structures
| Homology group | Gene | Structure |
S. pneumoniae serotype(s) possessing gene |
|||
|---|---|---|---|---|---|---|
| Donor | Linkage | Acceptor | CPS structure known | CPS structure unknown | ||
| 81 | wciO | Ribitol | 5-P-2 | β-Galf | 6A, 6B, 33B, 33D | |
| 46 | wcrO | Ribitol | 5-P-3 | β-Galf | 33C, 34 | 35F, 36 |
| 87 | wcrB | Ribitol | 5-P-5 | β-Galf | 10F, 10A, 10B, 10C | |
| 149 | whaI | Ribitol | 5-P-6 | β-Galf | 39, 47F, 47A | 43 |
| 24 | wefM | Galp | α1-1 | Ribitol-5-P | 39 | |
| 24 | wcrC | Galp | α1-2 | Ribitol-5-P | 10A, 10C, 34, 47F, 47A | 35F, 43 |
| 24 | wcrF | Galp | α1-4 | Ribitol-5-P | 10B, 10F, 33B, 33D | |
| 25 | wciF | GalNAc | β1-3 | α-Gal | 10F, 10A, 10B, 10C, 33B, 33C, 33D, 39 | 13, 36 |
| 32 | wcrD | Galf | β1-3 | β-GalNAc | 10F,a 10A, 10B, 10C,a 33C, 34, 39, 47F | 13, 35F |
| 102 | wcrG | Galp | β1-6 | β-GalNAc | 10A, 10B,a 39 | |
The gene of interest occurs as a pseudogene in this serotype (9).
Whereas the four putative ribitol-5-phosphate transferases considered above are all members of different homology groups, the glycosyltransferases that act subsequently to link α-Galp to different positions of ribitol-5-phosphate all belong to the same homology group. Genes in this group were initially designated wcrC (6) and associated with the α1-2 transfer of Galp to ribitol-5-phosphate, based on the well-established structures of CPS10A and CPS34 (6). This association now extends to CPS10C (9), CPS47F (Fig. 4B), and CPS47A (19), and we expect it will also hold for CPS35F and CPS43, based on the relatively high homology that exists between genes designated wcrC in these serotypes. In contrast, the wcrC homologues in serotypes 10F (10) and 10B (9), as well as those in serotypes 33B and 33D (17), are associated with α1-4 (rather than α1-2) linkages between Galp and ribitol-5-phosphate and, thus, have been renamed wcrF (10) in Table 2. In contrast with these serotypes, the wcrC homologue of S. pneumoniae serotype 39 is associated with an α1-1 linkage between Galp and ribitol-5-phosphate, the same linkage that was associated with wefM of S. oralis (8). In accordance with these findings and the relatively high homology noted between these genes across the region (i.e., 75% identity across the first 130 amino acid residues) associated with linkage specificity (8), the wcrC homologue of S. pneumoniae serotype 39 has been renamed wefM in both Table 2 and Fig. 4. Of the 13 wcrC homologues that occur in different S. pneumoniae serotypes, the only one not listed in Table 2 is that of CPS serotype 33C, which was recently associated with α1-3 linkages between Galp and ribitol-5-phosphate, based on the results of GC-MS analysis of O-methylalditol acetates (17). Confirmation of this linkage by NMR would indicate the existence of a fourth gene for the transfer of α-Galp to ribitol-5-phosphate.
We associated wciF with the β1-3 transfer of GalNAc to Galp in the synthesis of CPS39 (Fig. 4B) based on the well-established role of this gene (and of the homologous wefD) in the synthesis of different S. pneumoniae CPS10 and Streptococcus gordonii or S. oralis RPS serotypes (6, 8, 9, 20, 21). However, recent studies of S. pneumoniae CPS serogroup 33 have been interpreted to suggest that wciF accounts for the α1-2 transfer of Galp to Galp (17). This interpretation is not compatible with our findings referenced above or with the predicted inverting mechanism of WciF (6). Instead, we suggest that genes designated wciF in CPS serogroup 33 are associated with β1-3 transfer of GalNAc to Galp in the synthesis of CPS33B, CPS33D, and CPS33C (Table 2) and with β1-3 transfer of Galp to Galp in the synthesis of CPS33A and CPS33F. Indeed, related β-GalNAc and β-Gal transferases also function in the synthesis of S. oralis RPS (20, 21). Based on available findings, the 13 wciF homologues in different S. pneumoniae CPS serotypes (5) appear to include 10 that are or may be associated with the β1-3 transfer of GalNAc (Table 2) and 2 (i.e., serotypes 33F and 33A) that are associated with the β1-3 transfer of Galp. The remaining wciF homologue occurs in the defective cps33F locus of S. pneumoniae serotype 37 (5) and has a coding sequence that is identical to those of the genes in S. pneumoniae serotypes 33F and 33A.
We previously showed by carbohydrate engineering (9, 10) that the Galf and Galp branches in CPS10A depend on wcrD, wciF, wcrG, and a member of wzy homology group 2 (wzy2) (Fig. 4A). We also predicted that the same genes in the cps39 locus were associated with similar braches in CPS39 and that these branches accounted for the cross-reaction of this serotype with CPS10A. The structure of CPS39 determined here confirms these predictions and thereby supports the proposed pathway for the formation of both branches. The critical steps are (i) wcrG-dependent transfer of β-Galp to subterminal β-GalNAc, which forms Galp branches in lipid-linked repeating subunits, and (ii) wzy2-dependent linkage of oligosaccharide repeating units through subterminal β-GalNAc, which forms Galf branches along the polysaccharide chain. Accordingly, the linear structures of CPS47F and CPS34 can be attributed to the absence of wcrG in the cps loci of these serotypes and the presence of polymerases (Wzy1 and Wzy31) that link oligosaccharide repeating subunits through terminal, wcrD-dependent Galf, which positions this residue in linear polysaccharide backbones (Fig. 4B).
The patterns of O-acetylation seen in CPS39, CPS47F, and CPS34 correlate well with the occurrence of wcyO, wciG, and wcjE for different putative O-acetyl transferases in the cps loci of these serotypes (Fig. 4A). The association of WcyO with 6-O-acetylation of wcrD-dependent Galf is evident both from the structures of CPS39 and CPS34 (Fig. 4B) and from the recently determined structure of CPS33C (17). Our assignment of WciG and WcjE to the two positions of O-acetylation of CPS47F (Fig. 4) was made in view of previous findings (17, 19, 22–24) that associate wciG with 2-O-acetylation of wcrD-dependent Galf in CPS serotypes 20B, 33F, 33A, 33B, 33D, and 35A and wcjE with 5,6-O-acetylation of wciB-dependent Galf in CPS serotypes 20, 33A, and 35A and 3,5-O-acetylation of 47A. The synthesis of S. pneumoniae CPS serogroup 35 involves a number of genes that are considered in the present study, as well as other genes (6, 7) that have not been associated with specific steps in polysaccharide biosynthesis. Comparative characterization of this serogroup is under way to gain further insights into the genetic basis of S. pneumoniae CPS structure and antigenicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Parastoo Azadi of CCRC, University of Georgia, for helpful discussions regarding chemical analyses of carbohydrate composition.
This work was supported in part by the Intramural Research Program of the NIDCR, NIH, and by NIH P01-HL-107153.
Footnotes
Published ahead of print 7 July 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01731-14.
REFERENCES
- 1.Pilishvili T, Noggel B, Moore MR. 2012. Chapter 11. Pneumococcal disease. In Roush SW, McIntyre L, Baldy LM. (ed), Manual for the surveillance of vaccine-preventable diseases, 5th ed. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 2.Lund E, Henrichsen J. 1978. Laboratory diagnosis, serology and epidemiology of Streptococcus pneumoniae, p 241–262 In Bergan T, Norris JR. (ed), Methods in microbiology. Academic Press, London, England [Google Scholar]
- 3.Garcia E, Llull D, Munoz R, Mollerach M, Lopez R. 2000. Current trends in capsular polysaccharide biosynthesis of Streptococcus pneumoniae. Res. Microbiol. 151:429–435. 10.1016/S0923-2508(00)00173-X [DOI] [PubMed] [Google Scholar]
- 4.Yother J. 2004. Capsules, p 30–48 In Tuomanen EI. (ed), The pneumococcus. ASM Press, Washington, DC [Google Scholar]
- 5.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. 10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. 2007. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7856–7876. 10.1128/JB.00837-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, Reeves PR, Bentley SD, Spratt BG. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7841–7855. 10.1128/JB.00836-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, Ritchey M, Yoshida Y, Bush CA, Cisar JO. 2009. Comparative structural and molecular characterization of ribitol-5-phosphate-containing Streptococcus oralis coaggregation receptor polysaccharides. J. Bacteriol. 191:1891–1900. 10.1128/JB.01532-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Nahm MH, Bush CA, Cisar JO. 2011. Comparative structural and molecular characterization of Streptococcus pneumoniae capsular polysaccharide serogroup 10. J. Biol. Chem. 286:35813–35822. 10.1074/jbc.M111.255422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Shelat NY, Bush CA, Cisar JO. 2010. Structure and molecular characterization of Streptococcus pneumoniae capsular polysaccharide 10F by carbohydrate engineering in Streptococcus oralis. J. Biol. Chem. 285:24217–24227. 10.1074/jbc.M110.123562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chittenden GJ, Roberts WK, Buchanan JG, Baddiley J. 1968. The specific substance from Pneumococcus type 34 (41). The phosphodiester linkages. Biochem. J. 109:597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, Roberts WK, Mills GT, Buchanan JG, Baddiley J. 1968. The O-acetyl groups of the specific substance from Pneumococcus type 34 (U.S. type 41). Carbohydr. Res. 8:262–265. 10.1016/S0008-6215(00)82231-4 [DOI] [Google Scholar]
- 13.Roberts WK, Buchanan JG, Baddiley J. 1963. The specific substance from Pneumococcus type 34 (41). The structure of a phosphorus-free repeating unit. Biochem. J. 88:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy N, Glaudemans CPJ. 1968. The specific substance from Diplococcus pneumoniae type 34 (u.s. type 41): the location of the O-acetyl groups. Carbohydr. Res. 8:214–218. 10.1016/S0008-6215(00)80157-3 [DOI] [Google Scholar]
- 15.Abeygunawardana C, Bush CA. 1993. Determination of the chemical structure of complex polysaccharides by heteronuclear NMR spectroscopy, p 199–249 In Bush CA. (ed), Advances in biophysical chemistry, vol 3 JAI Press, Greenwich, CT [Google Scholar]
- 16.Kamerling JP. 2000. Pneumococcal polysaccharides: a chemical view, p 81–114 In Tomasz A. (ed), Streptococcus pneumoniae: molecular biology & mechanisms of disease. Mary Ann Liebert, Inc., Larchmont, NY [Google Scholar]
- 17.Lin FL, Vinogradov E, Deng C, Zeller S, Phelan L, Green BA, Jansen KU, Pavliak V. 2014. Structure elucidation of capsular polysaccharides from Streptococcus pneumoniae serotype 33C, 33D, and revised structure of serotype 33B. Carbohydr. Res. 383:97–104. 10.1016/j.carres.2013.11.006 [DOI] [PubMed] [Google Scholar]
- 18.Jones C. 1995. Full assignment of the NMR spectrum of the capsular polysaccharide from Streptococcus pneumoniae serotype 10A. Carbohydr. Res. 269:175–181. 10.1016/0008-6215(94)00340-L [DOI] [PubMed] [Google Scholar]
- 19.Petersen BO, Hindsgaul O, Paulsen BS, Redondo AR, Skovsted IC. 2014. Structural elucidation of the capsular polysaccharide from Streptococcus pneumoniae serotype 47A by NMR spectroscopy. Carbohydr. Res. 386:62–67. 10.1016/j.carres.2013.11.013 [DOI] [PubMed] [Google Scholar]
- 20.Yoshida Y, Ganguly S, Bush CA, Cisar JO. 2005. Carbohydrate engineering of the recognition motifs in streptococcal coaggregation receptor polysaccharides. Mol. Microbiol. 58:244–256. 10.1111/j.1365-2958.2005.04820.x [DOI] [PubMed] [Google Scholar]
- 21.Yoshida Y, Yang J, Peaker PE, Kato H, Bush CA, Cisar JO. 2008. Molecular and antigenic characterization of a Streptococcus oralis coaggregation receptor polysaccharide by carbohydrate engineering in Streptococcus gordonii. J. Biol. Chem. 283:12654–12664. 10.1074/jbc.M801412200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calix JJ, Porambo RJ, Brady AM, Larson TR, Yother J, Abeygunwardana C, Nahm MH. 2012. Biochemical, genetic, and serological characterization of two capsule subtypes among Streptococcus pneumoniae serotype 20 strains: discovery of a new pneumococcal serotype. J. Biol. Chem. 287:27885–27894. 10.1074/jbc.M112.380451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calix JJ, Saad JS, Brady AM, Nahm MH. 2012. Structural characterization of Streptococcus pneumoniae serotype 9A capsule polysaccharide reveals role of glycosyl 6-O-acetyltransferase wcjE in serotype 9V capsule biosynthesis and immunogenicity. J. Biol. Chem. 287:13996–14003. 10.1074/jbc.M112.346924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemercinier X, Jones C. 2006. Full assignment of the 1H and 13C spectra and revision of the O-acetylation site of the capsular polysaccharide of Streptococcus pneumoniae type 33F, a component of the current pneumococcal polysaccharide vaccine. Carbohydr. Res. 341:68–74. 10.1016/j.carres.2005.10.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.