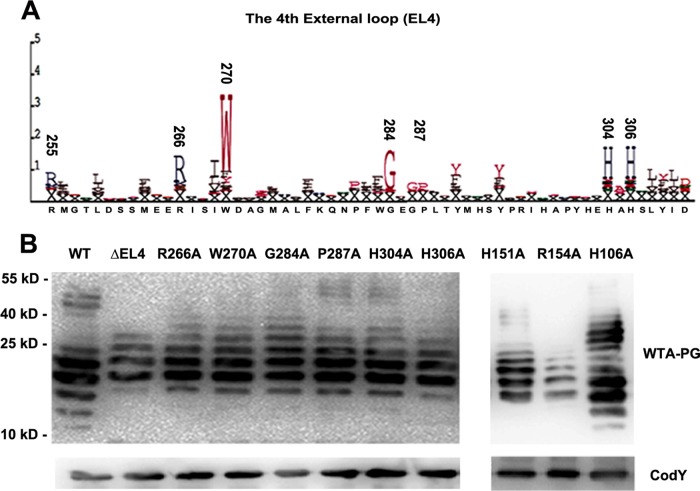
FIG 3.
Amino acid substitutions of the predicted conserved residues and deletion of EL4 impair the function of RafX. (A) Representation of conserved amino acids in EL4 generated by the SAM-T02 protein structure prediction server. The positions of the most-conserved residues are indicated above the letters. The size of each letter is proportional to the level of conservation of the amino acid. (B) Contributions of the predicted EL4 domain and conserved key amino acid residues to the WTA-PG banding pattern. Conserved residues R266, W270, G284A, P287A, H304, and H306, as well as H151 and R154 in the second external loop, were replaced with alanines to prepare ΔrafX mutant D39, and H106A (internal loop) served as a predicted negative control. Samples from the mutants were subjected to immunoblotting with antibodies specific for P-Cho and CodY. CodY was used as a loading control. The positions of size markers (molecular masses in kilodaltons are shown) are indicated.