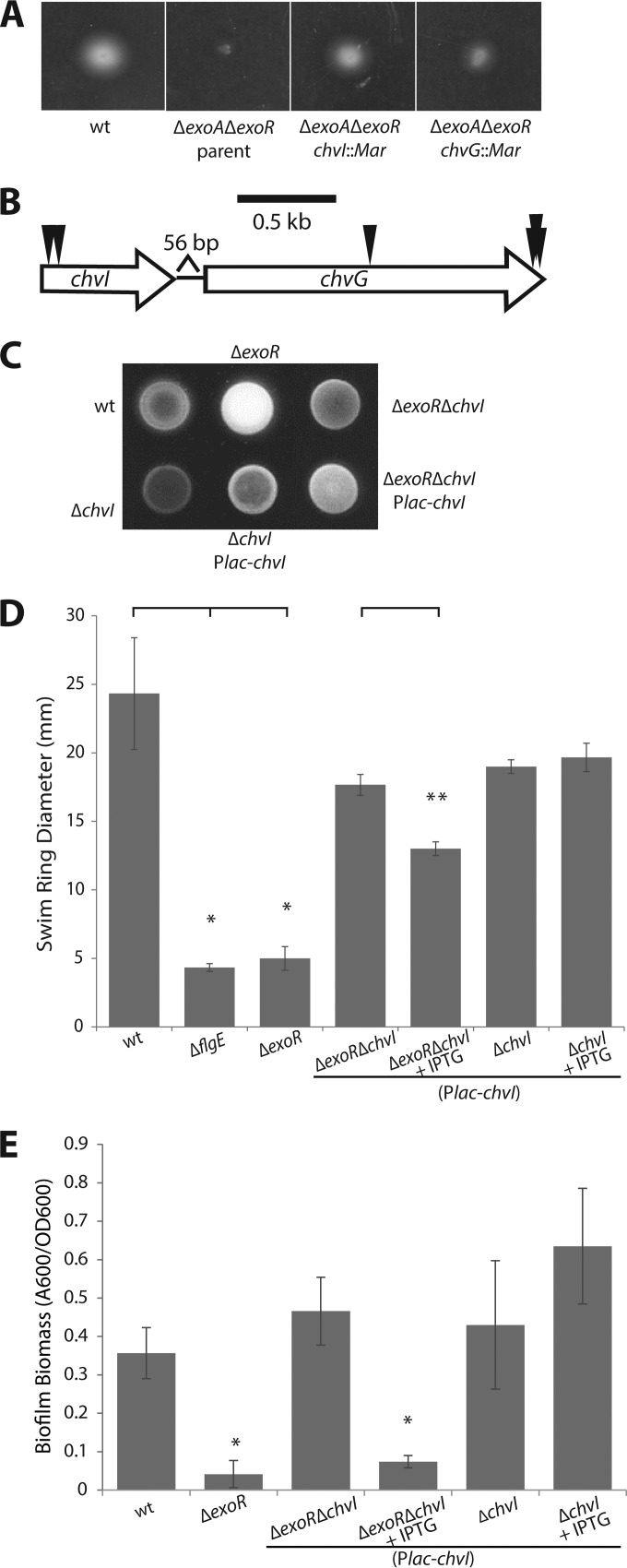
FIG 2.
Activity of the two-component system ChvG-ChvI directs exoR phenotypes. (A) Image of swim plate 3 days postinoculation of the parent ΔexoA ΔexoR strain and suppressor transposon mutants in chvI and chvG. (B) Genomic organization of the chvI-chvG locus and positions of independent transposon insertions. (C) Calcofluor white fluorescence under UV light exposure as a qualitative representation of the level of SCG biosynthesis. (D) Quantification of flagellar locomotion through swim agar (0.3%) 3 days postinoculation. Single asterisks denote a significant difference relative to the wild type (P < 0.05 with Bonferroni's correction). Double asterisks denote a significant difference between ΔexoR ΔchvI (plac-chvI) strains without and with IPTG (P = 0.009). Error bars represent standard deviations of the means of three biological replicates. (E) Biofilm formation on PVC coverslips in static culture. The A600/OD600 unit represents biofilm biomass as measured by solubilized crystal violet staining of biofilms formed on coverslips normalized to culture density. Asterisks denote a significant difference (P < 0.05 with Bonferroni's correction) compared to the wild type. Error bars represent standard deviations of the means of three biological replicates of a representative experiment.