TABLE 1.
Run time on different data sizesa
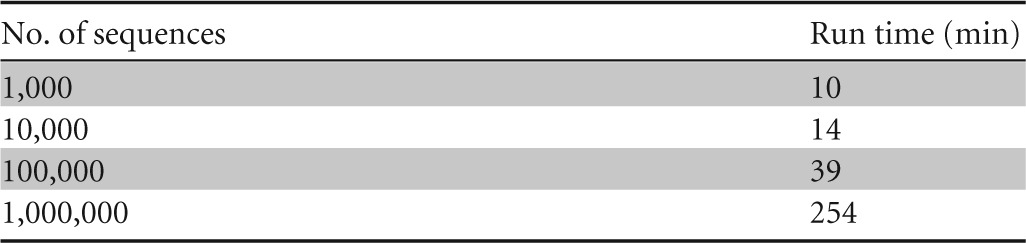
For these experiments, we used a MacBookPro 7.1 with a 2.66-GHz Intel Core 2 Duo processor and 8 GB 1067-MHz DDR RAM. Aligned bacterial 16S sequences from RDP (v.10.18) were downloaded in fasta format and trimmed using TrimAlignment such that only the 1,542 alignment columns with nucleotides in the Escherichia coli sequence with RDP code S001099426 were kept. Sequences shorter than 1,000 bp (excluding gaps) were removed. From the remaining 1.1 million sequences, random subsets of 1,000, 10,000, 100,000, and 1,000,000 sequences were extracted. DegePrime was run on these subsets using dmax = 12 and l = 18.
