FIG 5.
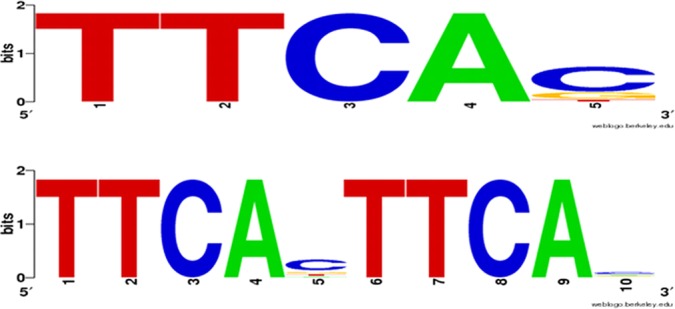
cre-tag analysis. The putative CreBC motif was found by analyzing the 500 bp upstream of differentially CreBC-regulated and CreBC-β-lactam-regulated genes/operons (P value, ≤0.01, ≥2-fold) (Tables 4 and 5) in PAOΔcreBC. The Pseudomonas Genome Database (35, 36) was used to identify the computationally predicted operons and the promoter-proximal TTCACnnnnnnTTCAC cre-tag motifs. The output is represented using WebLogo (53) for the creBC-dependent genes, including the presence of the single tag (top) and the direct double repeats (bottom).
