Abstract
An experimental assay was developed to search for proteins capable of antagonizing histone H1-mediated general repression of transcription. T7 RNA polymerase templates containing an upstream scaffold-associated region (SAR) were highly selectively repressed by H1 relative to non-SAR control templates. This is due to the nucleation of H1 assembly into flanking DNA brought about by the numerous A-tracts (AT-rich sequences containing short homopolymeric runs of dA.dT base pairs) of the SAR. Partial, selective titration of these A-tracts by the high mobility group (HMG) protein HMG-I/Y led to the complete derepression of transcription from the SAR template by inducing the redistribution of H1 on to non-SAR templates. SARs are associated with many highly transcribed regulated genes where they may serve to facilitate the HMG-I/Y-mediated displacement of histone H1 in chromatin. Indeed, HMG-I/Y was found to be strongly enriched in the H1-depleted subfraction which can be isolated from chromatin.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Käs E., Laemmli U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989 Dec 20;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y., Luke M., Laemmli U. K. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991 Jan 11;64(1):137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Albanese I., Weintraub H. Electrophoretic separation of a class of nucleosomes enriched in HMG 14 and 17 and actively transcribed globin genes. Nucleic Acids Res. 1980 Jun 25;8(12):2787–2805. doi: 10.1093/nar/8.12.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasquez V. C., Xu M., Moses S. C., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. I. Role of the intronic MAR and enhancer in plasmacytoma cells. J Biol Chem. 1989 Dec 15;264(35):21183–21189. [PubMed] [Google Scholar]
- Bode J., Maass K. Chromatin domain surrounding the human interferon-beta gene as defined by scaffold-attached regions. Biochemistry. 1988 Jun 28;27(13):4706–4711. doi: 10.1021/bi00413a019. [DOI] [PubMed] [Google Scholar]
- Chambers S. A., Rill R. L. Enrichment of transcribed and newly replicated DNA in soluble chromatin released from nuclei by mild micrococcal nuclease digestion. Biochim Biophys Acta. 1984 Jun 16;782(2):202–209. doi: 10.1016/0167-4781(84)90025-3. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Travers A. A. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991 Mar;16(3):92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Yuen M. H., Garrard W. T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987 Apr 15;262(11):5394–5397. [PubMed] [Google Scholar]
- Croston G. E., Kerrigan L. A., Lira L. M., Marshak D. R., Kadonaga J. T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991 Feb 8;251(4994):643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- De Bernardin W., Losa R., Koller T. Formation and characterization of soluble complexes of histone H1 with supercoiled DNA. J Mol Biol. 1986 Jun 5;189(3):503–517. doi: 10.1016/0022-2836(86)90320-7. [DOI] [PubMed] [Google Scholar]
- Dickinson L. A., Joh T., Kohwi Y., Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992 Aug 21;70(4):631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Eckner R., Birnstiel M. L. Cloning of cDNAs coding for human HMG I and HMG Y proteins: both are capable of binding to the octamer sequence motif. Nucleic Acids Res. 1989 Aug 11;17(15):5947–5959. doi: 10.1093/nar/17.15.5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton T. S., Reeves R. Purification and postsynthetic modifications of Friend erythroleukemic cell high mobility group protein HMG-I. Anal Biochem. 1986 Aug 15;157(1):53–62. doi: 10.1016/0003-2697(86)90195-8. [DOI] [PubMed] [Google Scholar]
- Ericsson C., Grossbach U., Björkroth B., Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990 Jan 12;60(1):73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin as an essential part of the transcriptional mechanism. Nature. 1992 Jan 16;355(6357):219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Garrard W. T. Histone H1 and the conformation of transcriptionally active chromatin. Bioessays. 1991 Feb;13(2):87–88. doi: 10.1002/bies.950130208. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Mathew C. G., Wright C. A., Venkov C. D., Johns E. W. Analysis of the high mobility group proteins associated with salt-soluble nucleosomes. Nucleic Acids Res. 1979 Dec 11;7(7):1815–1835. doi: 10.1093/nar/7.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Homberger H. P. Bent DNA is a structural feature of scaffold-attached regions in Drosophila melanogaster interphase nuclei. Chromosoma. 1989 Aug;98(2):99–104. doi: 10.1007/BF00291044. [DOI] [PubMed] [Google Scholar]
- Huang S. Y., Garrard W. T. Electrophoretic analyses of nucleosomes and other protein-DNA complexes. Methods Enzymol. 1989;170:116–142. doi: 10.1016/0076-6879(89)70044-6. [DOI] [PubMed] [Google Scholar]
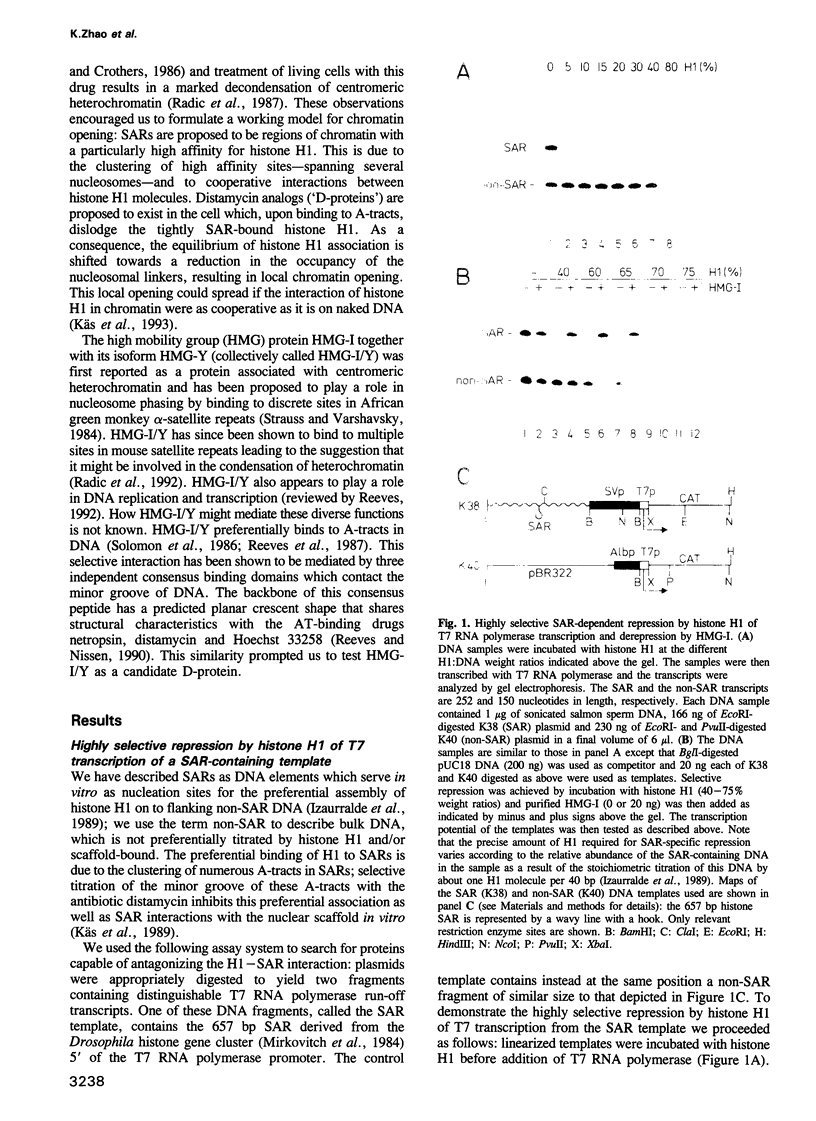
- Izaurralde E., Käs E., Laemmli U. K. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J Mol Biol. 1989 Dec 5;210(3):573–585. doi: 10.1016/0022-2836(89)90133-2. [DOI] [PubMed] [Google Scholar]
- Jackson J. B., Pollock J. M., Jr, Rill R. L. Chromatin fractionation procedure that yields nucleosomes containing near-stoichiometric amounts of high mobility group nonhistone chromosomal proteins. Biochemistry. 1979 Aug 21;18(17):3739–3748. doi: 10.1021/bi00584a015. [DOI] [PubMed] [Google Scholar]
- Jarman A. P., Higgs D. R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988 Nov;7(11):3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Disney J. E., Wyatt C. R., Reeves R. Expression of mRNAs encoding mammalian chromosomal proteins HMG-I and HMG-Y during cellular proliferation. Exp Cell Res. 1990 Mar;187(1):69–76. doi: 10.1016/0014-4827(90)90118-t. [DOI] [PubMed] [Google Scholar]
- Johnson K. R., Lehn D. A., Elton T. S., Barr P. J., Reeves R. Complete murine cDNA sequence, genomic structure, and tissue expression of the high mobility group protein HMG-I(Y). J Biol Chem. 1988 Dec 5;263(34):18338–18342. [PubMed] [Google Scholar]
- Johnson K. R., Lehn D. A., Reeves R. Alternative processing of mRNAs encoding mammalian chromosomal high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol. 1989 May;9(5):2114–2123. doi: 10.1128/mcb.9.5.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost J. P., Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Thomas J. O. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990 Dec;9(12):3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klehr D., Maass K., Bode J. Scaffold-attached regions from the human interferon beta domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry. 1991 Feb 5;30(5):1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U. K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J Mol Biol. 1989 Dec 5;210(3):587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- Käs E., Laemmli U. K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992 Feb;11(2):705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käs E., Poljak L., Adachi Y., Laemmli U. K. A model for chromatin opening: stimulation of topoisomerase II and restriction enzyme cleavage of chromatin by distamycin. EMBO J. 1993 Jan;12(1):115–126. doi: 10.1002/j.1460-2075.1993.tb05637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K., Käs E., Poljak L., Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992 Apr;2(2):275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Loc P. V., Strätling W. H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988 Mar;7(3):655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus M. E., de Graaf A., Mattia E., den Blaauwen J. L., Grande M. A., de Jong L., van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992 Sep 18;70(6):949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- Lund T., Dahl K. H., Mørk E., Holtlund J., Laland S. G. The human chromosomal protein HMG I contains two identical palindrome amino acid sequences. Biochem Biophys Res Commun. 1987 Jul 31;146(2):725–730. doi: 10.1016/0006-291x(87)90589-4. [DOI] [PubMed] [Google Scholar]
- Mathew C. G., Goodwin G. H., Johns E. W. Studies on the association of the high mobility group non-histone chromatin proteins with isolated nucleosomes. Nucleic Acids Res. 1979 Jan;6(1):167–179. doi: 10.1093/nar/6.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mueller C. R., Maire P., Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990 Apr 20;61(2):279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- Nissen M. S., Langan T. A., Reeves R. Phosphorylation by cdc2 kinase modulates DNA binding activity of high mobility group I nonhistone chromatin protein. J Biol Chem. 1991 Oct 25;266(30):19945–19952. [PubMed] [Google Scholar]
- Palvimo J., Linnala-Kankkunen A. Identification of sites on chromosomal protein HMG-I phosphorylated by casein kinase II. FEBS Lett. 1989 Oct 23;257(1):101–104. doi: 10.1016/0014-5793(89)81796-x. [DOI] [PubMed] [Google Scholar]
- Phi-Van L., von Kries J. P., Ostertag W., Strätling W. H. The chicken lysozyme 5' matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990 May;10(5):2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Radic M. Z., Lundgren K., Hamkalo B. A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987 Sep 25;50(7):1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- Radic M. Z., Saghbini M., Elton T. S., Reeves R., Hamkalo B. A. Hoechst 33258, distamycin A, and high mobility group protein I (HMG-I) compete for binding to mouse satellite DNA. Chromosoma. 1992 Oct;101(10):602–608. doi: 10.1007/BF00360537. [DOI] [PubMed] [Google Scholar]
- Reeves R. Chromatin changes during the cell cycle. Curr Opin Cell Biol. 1992 Jun;4(3):413–423. doi: 10.1016/0955-0674(92)90006-x. [DOI] [PubMed] [Google Scholar]
- Reeves R., Elton T. S., Nissen M. S., Lehn D., Johnson K. R. Posttranscriptional gene regulation and specific binding of the nonhistone protein HMG-I by the 3' untranslated region of bovine interleukin 2 cDNA. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6531–6535. doi: 10.1073/pnas.84.18.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Langan T. A., Nissen M. S. Phosphorylation of the DNA-binding domain of nonhistone high-mobility group I protein by cdc2 kinase: reduction of binding affinity. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1671–1675. doi: 10.1073/pnas.88.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R., Nissen M. S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990 May 25;265(15):8573–8582. [PubMed] [Google Scholar]
- Romig H., Fackelmayer F. O., Renz A., Ramsperger U., Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 1992 Sep;11(9):3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Sandeen G., Wood W. I., Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980 Sep 11;8(17):3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A., Fascher K. D., Hörz W. Nucleosome disruption at the yeast PHO5 promoter upon PHO5 induction occurs in the absence of DNA replication. Cell. 1992 Nov 27;71(5):853–864. doi: 10.1016/0092-8674(92)90560-y. [DOI] [PubMed] [Google Scholar]
- Sen D., Crothers D. M. Influence of DNA-binding drugs on chromatin condensation. Biochemistry. 1986 Apr 8;25(7):1503–1509. doi: 10.1021/bi00355a005. [DOI] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Solomon M. J., Strauss F., Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A.T base pairs in duplex DNA. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stief A., Winter D. M., Strätling W. H., Sippel A. E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989 Sep 28;341(6240):343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Thanos D., Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992 Nov 27;71(5):777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- Uemura T., Ohkura H., Adachi Y., Morino K., Shiozaki K., Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987 Sep 11;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Vyas P., Vickers M. A., Simmons D. L., Ayyub H., Craddock C. F., Higgs D. R. Cis-acting sequences regulating expression of the human alpha-globin cluster lie within constitutively open chromatin. Cell. 1992 May 29;69(5):781–793. doi: 10.1016/0092-8674(92)90290-s. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992 Dec 11;258(5089):1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- Xu M., Hammer R. E., Blasquez V. C., Jones S. L., Garrard W. T. Immunoglobulin kappa gene expression after stable integration. II. Role of the intronic MAR and enhancer in transgenic mice. J Biol Chem. 1989 Dec 15;264(35):21190–21195. [PubMed] [Google Scholar]
- von Kries J. P., Buhrmester H., Strätling W. H. A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell. 1991 Jan 11;64(1):123–135. doi: 10.1016/0092-8674(91)90214-j. [DOI] [PubMed] [Google Scholar]