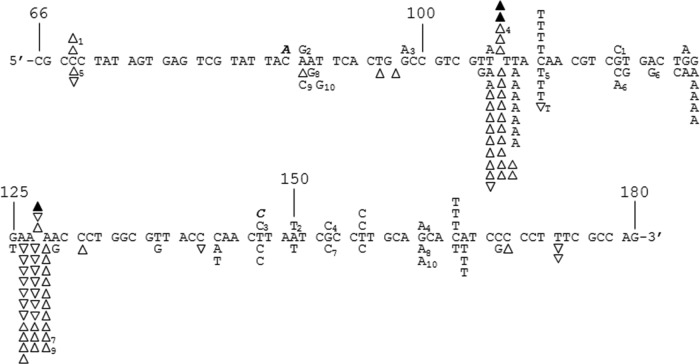
FIG 2.
DNA sequence analysis from the PCR-based lacZα complementation fidelity assay. The 115-base region analyzed for mutations is shown (see Fig. 1). The coding strand for lacZα is shown in the 5′-3′ direction (bottom strand in Fig. 1C). Numbering is as shown in Fig. 1C. Deletions are shown as regular triangles, insertions are shown as downward triangles with the inserted base shown adjacent to the downward triangle, unless it was same as the base in a nucleotide run, and base substitutions are shown directly above or below the sequence. Substitutions shown correspond to the recovered sequence for the coding strand; however, these mutations could have occurred during synthesis of the noncoding strand as well (i.e., a C-to-A change shown here could have resulted from a C-to-A change during synthesis of the coding strand or a G-to-T change during synthesis of the noncoding strand) (see Fig. 1 and Results). Mutations recovered from HIV RT at 6 mM Mg2+ and 100 μM dNTPs and mutations from background controls are shown above the sequence as open triangles and normal text or filled triangles and bold italicized text, respectively. Mutations from HIV RT at 0.25 mM Mg2+ and 5 μM dNTPs are shown below the sequence. Individual sequence clones which had multiple mutations (more than one mutation event) are marked with subscripts adjacent to the mutations. Several clones with deletions (either single or multiple deletions) at positions 181 to 183, just outside the scored region, were also recovered (data not shown). This was the dominant mutation type recovered in background controls (19 out of 24 total sequences) and probably resulted from improper ligation events or damaged plasmid vectors (see Results). Two out of 22 HIV RT-derived sequences at 6 mM Mg2+ and 62 out of 162 HIV RT-derived sequences at 0.25 mM Mg2+ also had these deletions.