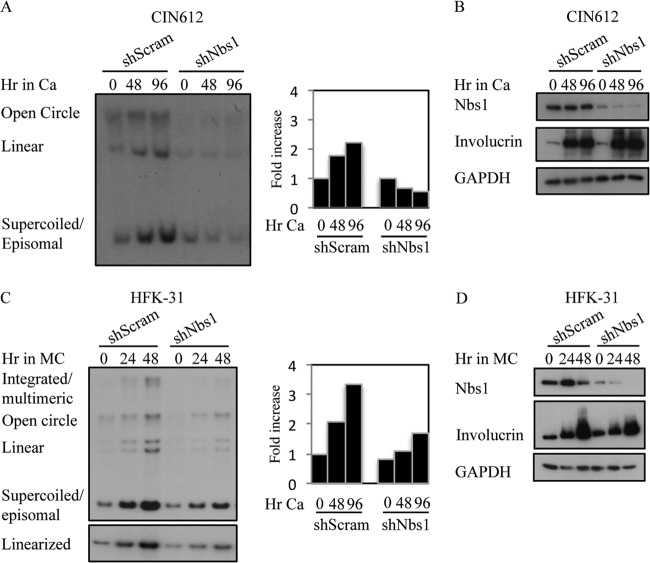
FIG 6.
Nbs1 is necessary for productive viral replication. (A) DNA was harvested from CIN612 9E cells stably expressing a Scramble shRNA or Nbs1 shRNA at T0 (undifferentiated) or after 48 and 96 h of differentiation in high-calcium medium. Southern blot analysis was performed to analyze viral genome amplification. The bar graph represents quantification of the episome copy number present at each time point, relative to T0 shScramble, which was set to 1. Densitometry was performed using ImageJ. Ca, calcium. (B) Total protein was harvested from CIN612 9E shScramble and shNbs1 cells at T0 or after 48 and 96 h of differentiation in high-calcium medium. Western blot analysis was performed using antibodies to Nbs1 and involucrin, and GAPDH as a loading control. (C) DNA was harvested from human foreskin keratinocytes stably maintaining HPV31 genomes (HFK-31) at T0 or after 24 and 48 h of differentiation in methylcellulose (MC) and analyzed by Southern blotting for amplification of viral genomes. DNA samples were digested with BamHI (does not cut the viral genome; upper panel) or with HindIII to linearize viral genomes. The bar graph represents quantification of the episome copy number present at each time point, relative to T0 shScramble (set to 1). Densitometry was performed using ImageJ. (D) Western blot analysis was performed on lysates harvested from HFK-31 cells at T0 or after 24 and 48 h of differentiation in methylcellulose using antibodies to Nbs1 and involucrin. GAPDH was used as a loading control. All results are representative of observations of four or more independent experiments.