ABSTRACT
Generalized immune activation during HIV infection is associated with an increased risk of cardiovascular disease, neurocognitive disease, osteoporosis, metabolic disorders, and physical frailty. The mechanisms driving this immune activation are poorly understood, particularly for individuals effectively treated with antiretroviral medications. We hypothesized that viral characteristics such as sequence diversity may play a role in driving HIV-associated immune activation. We therefore sequenced proviral DNA isolated from peripheral blood mononuclear cells from HIV-infected individuals on fully suppressive antiretroviral therapy. We performed phylogenetic analyses, calculated viral diversity and divergence in the env and pol genes, and determined coreceptor tropism and the frequency of drug resistance mutations. Comprehensive immune profiling included quantification of immune cell subsets, plasma cytokine levels, and intracellular signaling responses in T cells, B cells, and monocytes. These antiretroviral therapy-treated HIV-infected individuals exhibited a wide range of diversity and divergence in both env and pol genes. However, proviral diversity and divergence in env and pol, coreceptor tropism, and the level of drug resistance did not significantly correlate with markers of immune activation. A clinical history of virologic failure was also not significantly associated with levels of immune activation, indicating that a history of virologic failure does not inexorably lead to increased immune activation as long as suppressive antiretroviral medications are provided. Overall, this study demonstrates that latent viral diversity is unlikely to be a major driver of persistent HIV-associated immune activation.
IMPORTANCE Chronic immune activation, which is associated with cardiovascular disease, neurologic disease, and early aging, is likely to be a major driver of morbidity and mortality in HIV-infected individuals. Although treatment of HIV with antiretroviral medications decreases the level of immune activation, levels do not return to normal. The factors driving this persistent immune activation, particularly during effective treatment, are poorly understood. In this study, we investigated whether characteristics of the latent, integrated HIV provirus that persists during treatment are associated with immune activation. We found no relationship between latent viral characteristics and immune activation in treated individuals, indicating that qualities of the provirus are unlikely to be a major driver of persistent inflammation. We also found that individuals who had previously failed treatment but were currently effectively treated did not have significantly increased levels of immune activation, providing hope that past treatment failures do not have a lifelong “legacy” impact.
INTRODUCTION
Generalized immune activation is a hallmark of HIV-1 infection. In this state, a variety of immune cells show an increase in expression of activation, proliferation, and apoptotic markers, cellular turnover with aberrant cell cycle regulation, production of proinflammatory cytokines, and increased lymphoid tissue fibrosis (1–6). Immune activation is strongly associated with HIV-1 disease progression; for instance, T cell activation, as measured by expression of CD38 and HLA-DR, is more predictive of CD4+ T cell depletion and shorter survival than is the plasma viral load (7, 8). Furthermore, the level of immune activation early in HIV-1 infection as measured by CD8+ T cell activation predicts CD4+ T cell loss independently of plasma HIV-1 RNA levels (9). Suppression of viral replication with effective antiretroviral treatment (ART) reduces immune activation, but even effective ART regimens are unable to reduce the levels of immune activation in HIV-infected individuals to levels seen in healthy individuals (1). Since this inflammation is linked to poor health outcomes, including elevated risks of death, cardiovascular disease (CVD), neurocognitive impairment, osteoporosis, and frailty even in treated individuals (1, 2, 10–22), understanding the mechanisms behind immune activation may lead to new treatments to improve the quality and length of life for HIV-1-infected individuals.
Viral characteristics such as the presence of drug resistance mutations, coreceptor tropism, and the diversity of the proviral population are associated with HIV-1 disease progression (23–27). For example, individuals initially infected with a more diverse viral population undergo more rapid HIV-1 disease progression (23, 24). However, it is unclear whether these viral characteristics drive poor health outcomes by influencing levels of immune activation. Several studies suggest that in viremic individuals, the presence of drug resistance mutations is associated with lower levels of inflammation. For instance, the number of mutations conferring drug resistance was inversely correlated with levels of the proinflammatory cytokines interleukin 6 (IL-6), tumor necrosis factor alpha (TNF-α), and tumor necrosis factor receptor II (TNF-rII), independent of HIV-1 RNA levels (28). In addition, individuals with documented drug resistance have lower levels of CD4+ and CD8+ T cell activation, experience lower rates of CD4+ depletion, and progress more slowly to AIDS than do untreated HIV-infected individuals, independently of plasma HIV RNA levels (28–32). The reduced ability of drug-resistant viruses to contribute to disease progression may result from particular drug resistance mutations that impede viral replicative capacity, thus decreasing the activation of bystander T cells (29, 33). Conversely, the presence of CXCR4-tropic viruses was associated with higher levels of inflammatory markers in one study (34) but with no difference in levels of inflammatory markers in another (35). However, few studies have evaluated associations between viral characteristics and inflammation.
Improvements in ART regimens now allow for the majority of HIV-infected patients under care to maintain virologic suppression (36). Understanding the mechanisms underlying HIV-associated chronic inflammation in treated disease—including the role of the latent proviral reservoir that is maintained despite effective treatment—could inform the design of ART strategies and the development of novel therapeutic strategies to reverse chronic inflammation and improve health outcomes. Several studies suggest that low-level HIV-1 replication in the plasma may persist despite suppressive treatment (37–42). In fact, approximately 75% of individuals who have “suppressed” HIV-1 RNA levels using standard assays have detectable viremia with an ultrasensitive single-copy assay (43). While two studies suggest that this low-level viral replication may drive immune activation in treated individuals (44, 45), in another study no such association was noted (46). Overall, the existing data seem to indicate that low-level plasma viremia may explain some, but not all, of the persistent immune activation. The proviral population in HIV-infected individuals serves as an archive of circulating plasma viruses from throughout the course of infection (47–50) that does not undergo significant viral evolution once treatment has begun (51–55). Thus, examination of the proviral population provides the opportunity to examine whether certain viral characteristics, established early in infection, are associated with ongoing inflammation despite effective treatment. For instance, viral diversity is associated with disease progression in untreated individuals, but the extent to which a diverse proviral population is associated with ongoing inflammation remains unknown. We hypothesized that viral characteristics such as increased diversity, divergence (the average distance of an individual's sequences from their calculated most recent common ancestor [MRCA] as a measure of intrasubject evolution), and frequency of CXCR4-tropic viruses are associated with increased immune activation. To address this hypothesis, we undertook a uniquely comprehensive evaluation of viral characteristics and immune status in a cohort of individuals with currently undetectable viral loads on effective ART. We examined proviral characteristics, including diversity, divergence, ARV resistance, and coreceptor tropism. To assess immune status, we evaluated immune cell phenotype, serum cytokine levels, and immune cell function by phosphor-specific flow cytometry (56, 57). Since half of the subjects had a history of virologic failure (VF), defined as documented resistance as a result of therapy or as a viral load greater than or equal to 200 copies/ml after 6 or more months of therapy, we also examined whether a history of failed therapy was associated with a lasting impact on the immune status.
MATERIALS AND METHODS
Study population and sample collection.
Subjects were drawn from the Stanford HIV Aging Cohort (SHAC), an ongoing prospective clinical cohort of virologically suppressed HIV-infected individuals. Sixteen subjects, all with undetectable viral loads for at least 6 months prior to the beginning of the study, were selected. Eight subjects had a history of virologic failure requiring alterations in their ART regimens to maintain suppression. Blood samples were collected between June and August 2009, and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Ficoll-Paque). Samples were cryopreserved in 90% heat-inactivated fetal bovine serum (FBS) and 10% dimethyl sulfoxide (DMSO) and thawed for immune profiling and for characterization of the latent provirus.
Immune profiling.
Comprehensive immune profiling was performed at the Human Immune Monitoring Center at Stanford University (http://iti.stanford.edu/himc/flow-cytometry.html) as described previously (57). Briefly, cryopreserved PBMC samples were thawed and evaluated by flow cytometry to determine the distribution of immune cell subsets, including enumeration of T, T regulatory (Treg) cells, B cells, dendritic cells, monocytes, NK cells, differentiation into Th1, Th2, and Th17 cell subsets, and T cell activation status. Intracellular signaling responses in T cells, B cells, and monocytes were assessed by phospho-flow cytometry to evaluate pSTAT-1, pSTAT-3, pSTAT-5, pERK1/2, pp38, and pPLCγ2 levels in response to IL-2, IL-6, IL-7, IL-10, IL-21, alpha interferon (IFN-α), and IFN-γ as described previously (57). Finally, a human 51-plex Luminex immunoassay was used to assess cytokine levels in plasma from these subjects.
Proviral sequence determination.
For evaluation of HIV proviral DNA sequences, a single vial of PBMCs was thawed and DNA was extracted from 2 million to 6 million PBMCs using the QIAamp DNA minikit (Qiagen) according to the manufacturer's instructions. The open reading frame containing the region of env from V1 to V5 and the open reading frame containing the protease (PR) and reverse transcriptase (RT) regions of pol were amplified from subject proviral DNA with Taq DNA polymerase (Fisher Scientific) by nested PCR using the primers listed in Table S1 in the supplemental material, with ACH-2 proviral DNA (Qiagen) as a positive control and water and genomic DNA from HIV-uninfected individuals as negative controls. The env and pol PCR products were confirmed by agarose gel electrophoresis. The env and pol genes were amplified by limiting dilution PCR such that less that 50% of the reactions had a product to confirm single genome amplification. Unincorporated primers and deoxynucleoside triphosphates (dNTPs) were removed from amplified products in a single-step enzymatic reaction using ExoSAP-IT (Affymetrix) according to the manufacturer's instructions. The products were then sequenced with the BigDye Terminator v3.1 cycle sequencing kit (Invitrogen) and sent to Molecular Cloning Laboratories (MCLAB; South San Francisco, CA) for fragment analysis. The sequencing primers are detailed in Table S1 in the supplemental material. The sequence data were assembled and edited in Sequencher version 5.1 (Gene Codes Corporation, Ann Arbor, MI). Sequences were entered into the NCBI Basic Local Alignment Search Tool (BLAST), as well as into the NCBI Genotyping Tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi), to confirm their identity and the absence of any contamination. Sequences with mixed peaks were excluded.
Sequence and phylogenetic analyses.
The sequences of the V1 to V5 and PR-RT amplicons, along with the corresponding regions of the subtype A reference sequences 92ug037 (U51190.1) and Q23-17 (AF004885.1), the subtype B reference sequences HXB2 (K03455.1) and BK132 (AY173951.1), the subtype C reference sequences 92BR025 (U52953.1) and ETH2220 (U46016.1), the subtype D reference sequences 94UG114 (U88824.1) and ELI (K03454.1), and the subtype H reference sequence 90CF056.1 (AF005496) (all accession numbers are from GenBank), were manually aligned in Geneious version 6.0.6 (Biomatters Limited, Auckland, New Zealand) using a ClustalW multiple alignment with a gap open penalty of 15 and a gap extension penalty of 6.66. The Hypermut tool of the HIV database maintained by the Los Alamos National Laboratory (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html) was used to detect APOBEC-induced hypermutated sequences with a P value of less than 0.05, which were excluded from phylogenetic analyses. The PR-RT and the V1 to V5 sequences of HXB2 were used as the reference sequences for this analysis. In total, nine env sequences were removed for hypermutation, one each from subjects 81, 82, 83, 84, 86, 91, 93, 96, and 98. Six pol sequences were removed, from subjects 84 (2 sequences), 86 (1 sequence), 89 (2 sequences), and 91 (1 sequence). Neighbor-joining, bootstrapped (over 100 iterations) phylogenetic trees for pol and env were built in Geneious using the HKY genetic distance model, with the PR-RT and V1 to V5 sequences of 90CF056.1 used as the outgroup. The DIVEIN program (http://indra.mullins.microbiol.washington.edu/DIVEIN/diver.html) was used to determine pairwise diversity and divergence from the MRCA for both env and pol. Calculations were made with the HKY85 substitution model with a fixed transition-to-transversion ratio of 4.
Determination of coreceptor tropism.
Geno2pheno [coreceptor] version 2.5 (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php; Max-Planck-Institut Informatik, Germany) was used to determine coreceptor tropism of the V3 region of env, with a false-positive rate of 5%. The proviral population of each subject was assigned an aggregate label based on the tropism of the individual sequences: a population with only R5-tropic viruses was labeled R5, a population with only X4-tropic viruses was labeled X4, and a population with at least one X4-tropic virus was labeled dual.
Determination of drug resistance score.
The average drug resistance score for the pol sequences obtained from each subject was determined by the Stanford HIV Drug Resistance Database version 6.3.0 (http://hivdb.stanford.edu/index.html, Stanford University, Stanford, CA). Resistances to lopinavir/ritonavir (LPV/r), lamivudine (3TC) and abacavir (ABC), and efavirenz (EFV) were chosen to represent resistance to protease inhibitors (PIs), nucleoside reverse transcriptase inhibitors (NRTIs), and nonnucleoside reverse transcriptase inhibitors (NNRTIs), respectively, for subsequent analyses.
Statistical analysis.
Cytokine data were log transformed prior to analysis, and phospho-flow data were normalized and transformed as previously described (57). Spearman's rank correlation was applied to characterize the relationship between immune parameters and viral features. The Wilcoxon rank sum test was used to compare immune features between subjects with and without a history of virologic failure. As numerous features were involved in addressing each of these research questions, numerous corresponding hypotheses were tested. To account for the issue of multiple testing, we controlled the false-discovery rate to be no more than 10% for each of the two main research questions being addressed. Statistical analyses were performed by the statistical package R (http://www.r-project.org/) and Excel (Microsoft Corporation).
Nucleotide sequence accession numbers.
The pol sequences were deposited into GenBank with accession numbers KJ528594 through KJ528716. The env sequences were deposited into GenBank with accession numbers KJ528717 through KJ528870.
RESULTS
Characteristics of subjects.
The study cohort consisted of 16 HIV-infected individuals from the Stanford HIV Aging Cohort (SHAC), an ongoing prospective clinical cohort of virologically suppressed HIV-infected individuals (Table 1). Enrollment criteria included ART adherence and an undetectable HIV-1 viral load for at least 6 months. The subjects ranged in age from 30 to 78 years of age, with nadir CD4 counts from 0 to 584 CD4+ T cells/μl. The mean contemporaneous CD4 count was 653 cells/μl (range, 206 to 1,221 cells/μl), and the mean length of suppression was 38.5 months (range, ≥6 to 137 months). Although all subjects had viral loads below the limit of detection for at least 6 months at the time of the study, eight of the subjects had a clinical history of virologic failure (VF). The subjects with a history of VF tended to be older (mean age, 57 versus 45 years) and have a lower nadir CD4 count (208 versus 248 cells/μl) and current CD4 count (519 versus 786 cells/μl) (Table 1).
TABLE 1.
Subject demographics
| Group | IDa | Age (yr) | Nadir CD4 count (cells/μl) | Current CD4 count (cells/μl) | Length of suppression (mo) |
|---|---|---|---|---|---|
| No history of VF | 84 | 57 | 400 | 594 | ≥6 |
| 87 | 56 | 288 | 520 | 41 | |
| 88 | 40 | 3 | 1,221 | 137 | |
| 93 | 35 | 279 | 438 | 13 | |
| 95 | 59 | 189 | 618 | ≥6 | |
| 96 | 40 | 362 | 1,015 | 19 | |
| 99 | 37 | 220 | 805 | ≥6 | |
| 100 | 40 | 246 | 1,080 | 17 | |
| Mean | 46 | 248 | 786 | 31 | |
| History of VF | 81 | 54 | 413 | 469 | 13 |
| 82 | 56 | 584 | 898 | 87 | |
| 83 | 78 | 0 | 545 | 30 | |
| 86 | 70 | 140 | 452 | 36 | |
| 89 | 30 | 155 | 573 | 46 | |
| 91 | 38 | 138 | 474 | 70 | |
| 92 | 64 | 82 | 205 | 23 | |
| 98 | 67 | 150 | 539 | 66 | |
| Mean | 57 | 208 | 519 | 46 |
ID, identification.
Diversity and divergence in env sequences from virologically suppressed adults.
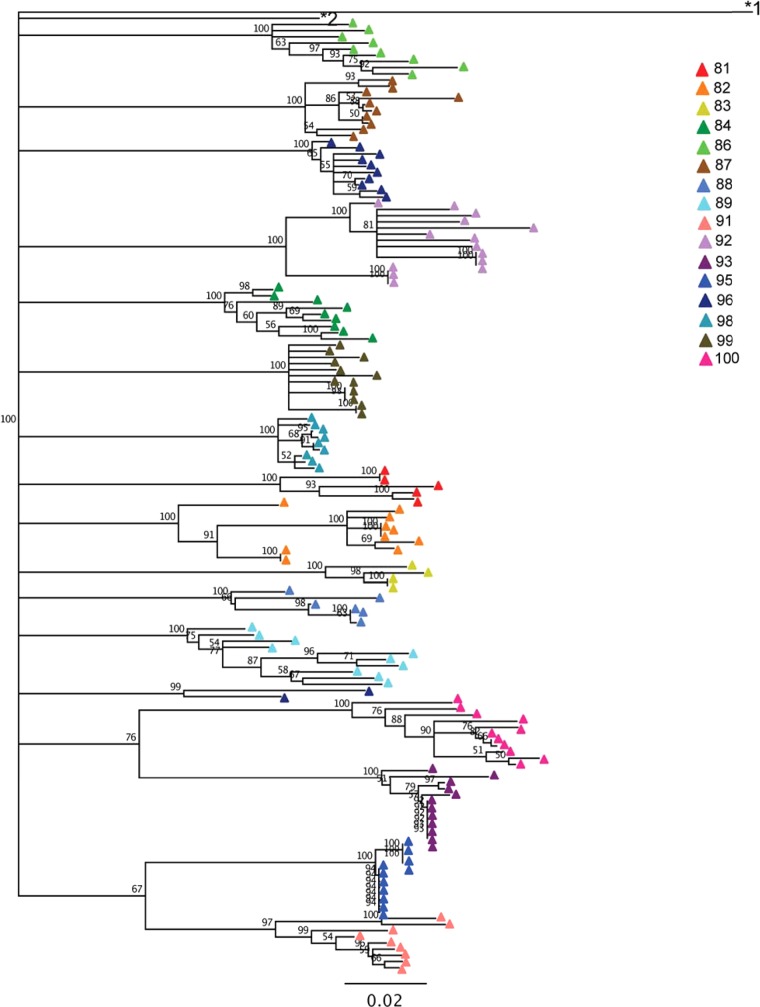
Neighbor-joining, bootstrapped phylogenetic trees of HIV-1 env V1 to V5 regions were used to quantify diversity and divergence (Fig. 1). Most subjects' sequences formed a monophyletic cluster, with the notable exception of subject 96, in whom two amplicons formed a unique cluster (Fig. 1). These two sequences did not cluster with any other subject, nor were they identified as known sequences by BLAST searching, indicating that subject 96 was most likely dually infected or superinfected with two unique strains from different partners. We were unable to obtain additional samples from this subject to confirm the dual infection. The subjects exhibited a wide range of diversity (0.35% to 5.0%) and divergence (6.8% to 13.0%) in their proviral populations (Table 2). These data indicate that a wide range of diversity in env sequences is observed even in individuals on effective therapy and with suppressed plasma viral loads.
FIG 1.
Neighbor-joining, bootstrapped phylogenetic tree of env sequences. Sequences are colored by subject. The numbers at the nodes indicate the percentage of bootstrap replicates (100 iterations total). The subtype H reference sequence 90CF056.1 (GenBank accession number AF005496) and subtype B reference sequence HXB2 (GenBank accession number K03455.1) are labeled *1 and *2, respectively.
TABLE 2.
env diversity and divergence and coreceptor tropisma
| ID | History of VF |
env diversity (%) |
env divergence (%) |
Coreceptor tropism | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| 84 | 0 | 3.52 | 1.07 | 8.18 | 0.89 | Dual |
| 87 | 0 | 2.53 | 1.19 | 9.78 | 0.56 | R5 |
| 88 | 0 | 2.98 | 2.07 | 8.55 | 0.83 | Dual |
| 93 | 0 | 1.28 | 1.24 | 11.11 | 0.37 | X4 |
| 95 | 0 | 0.35 | 0.36 | 10.96 | 0.32 | R5 |
| 96 | 0 | 4.54 | 4.99 | 9.89 | 0.56 | R5 |
| 99 | 0 | 1.71 | 0.69 | 9.13 | 0.51 | R5 |
| 100 | 0 | 3.66 | 1.36 | 12.98 | 0.71 | Dual |
| 81 | 1 | 4.52 | 2.15 | 9.72 | 0.55 | R5 |
| 82 | 1 | 3.70 | 2.50 | 9.19 | 1.44 | R5 |
| 83 | 1 | 2.61 | 1.58 | 9.85 | 0.45 | Dual |
| 86 | 1 | 3.98 | 1.20 | 6.78 | 1.02 | R5 |
| 89 | 1 | 5.00 | 1.37 | 8.74 | 1.88 | Dual |
| 91 | 1 | 3.91 | 2.60 | 10.80 | 0.95 | R5 |
| 92 | 1 | 4.45 | 2.05 | 12.03 | 1.23 | R5 |
| 98 | 1 | 1.08 | 0.32 | 7.47 | 0.20 | R5 |
| Mean | NA | 3.11 | 1.67 | 9.70 | 0.78 | NA |
ID, identification; VF, virologic failure (1 = true); NA, not available (we were unable to successfully obtain any amplicons).
Diversity, divergence, and drug resistance mutations in pol sequences from virologically suppressed adults.
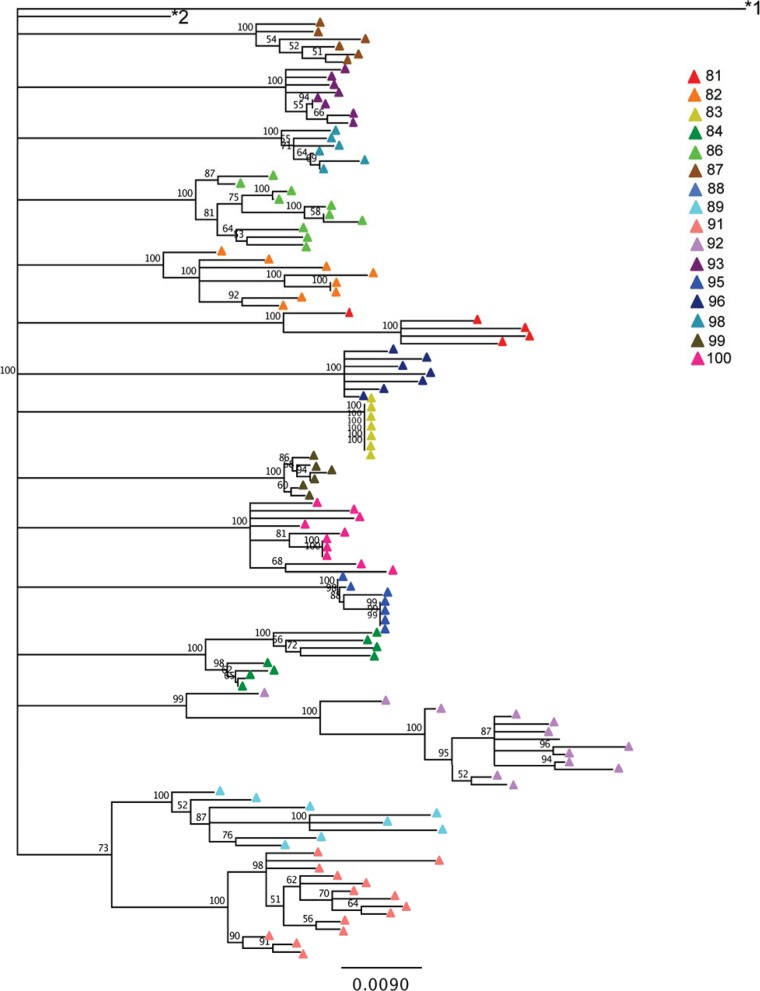
As with env, phylogenetic trees were constructed based on pol sequences in proviral DNA (Fig. 2). Amplification was unsuccessful for subject 88. The pol sequences exhibited less diversity and divergence than the env sequences, though a range in diversity (0% to 2.66%) and divergence (2.83% to 5.97%) was still observed (Table 3). Notably, subject 83 demonstrated no pol diversity (mean = 0; standard deviation [SD] = 0) or variability in divergence (mean = 4.2%; SD = 0%), indicating that all of the amplicon sequences were identical. Subject 83 had a nadir CD4+ T cell count of 0/μl, perhaps indicating that a population bottleneck substantially reduced the diversity of the proviral population. However, this phenomenon was not reflected for env, for which both diversity (mean = 2.61%; SD = 1.58%) and divergence (mean = 9.85%; SD = 0.45%) were observed (Table 2).
FIG 2.
Neighbor-joining, bootstrapped phylogenetic tree of pol sequences. Sequences are colored by subject. The subtype H reference sequence 90CF056.1 (GenBank accession number AF005496) and subtype B reference sequence HXB2 (GenBank accession number K03455.1) are labeled *1 and *2, respectively.
TABLE 3.
pol diversity and divergence and ART resistance scoresa
| ID | History of VF |
pol diversity (%) |
pol divergence (%) |
Mean resistance score for: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | LPV/r | 3TC | ABC | EFV | ||
| 84 | 0 | 1.82 | 0.72 | 3.39 | 0.72 | 0 | 3.13 | 16.25 | 0 |
| 87 | 0 | 1.33 | 0.42 | 4.04 | 0.23 | 0 | 0 | 0 | 0 |
| 88 | 0 | NA | NA | NA | NA | NA | NA | NA | NA |
| 93 | 0 | 0.78 | 0.26 | 3.59 | 0.19 | 0 | 1.88 | 1.88 | 0 |
| 95 | 0 | 0.43 | 0.35 | 4.58 | 0.23 | 0 | 2.86 | 2.86 | 0 |
| 96 | 0 | 0.91 | 0.24 | 4.89 | 0.26 | 0 | 0 | 10 | 0 |
| 99 | 0 | 0.41 | 0.12 | 3.62 | 0.08 | 0 | 0 | 0 | 3.33 |
| 100 | 0 | 1.50 | 0.63 | 3.69 | 0.31 | 1 | 0 | 0 | 1.5 |
| 81 | 1 | 2.46 | 0.61 | 5.78 | 0.91 | 0 | 60 | 35 | 70 |
| 82 | 1 | 2.07 | 0.66 | 3.71 | 0.56 | 1.25 | 33.13 | 18.75 | 0 |
| 83 | 1 | 0 | 0 | 4.20 | 0 | 0 | 75 | 75 | 0 |
| 86 | 1 | 1.71 | 0.61 | 3.16 | 0.40 | 0 | 29.5 | 26 | 10.5 |
| 89 | 1 | 2.66 | 0.86 | 3.77 | 0.96 | 0 | 13.57 | 22.86 | 12.86 |
| 91 | 1 | 1.67 | 0.60 | 4.37 | 0.58 | 0 | 33.93 | 40 | 6.43 |
| 92 | 1 | 2.16 | 1.34 | 5.97 | 1.23 | 57.69 | 78.85 | 76.13 | 51.92 |
| 98 | 1 | 0.82 | 0.35 | 2.83 | 0.24 | 0 | 0 | 0 | 0 |
| Mean | NA | 1.38 | 0.52 | 4.11 | 0.46 | 4.00 | 22.12 | 21.65 | 10.44 |
ID, identification; VF, virologic failure (1 = true); NA, not available (we were unable to successfully obtain any amplicons).
The Stanford HIV Drug Resistance Database, which produces a numeric score in addition to the discrete resistance categories for each drug, was used to evaluate the presence of drug resistance mutations in the proviral population for each subject. Scores for 3TC, ABC, EFV, and LPV/r were used to capture low- and high-level resistance to nucleoside reverse transcriptase inhibitors, nonnucleoside reverse transcriptase inhibitors, and protease inhibitors. ART resistance scores varied among the subjects, with the most resistance to 3TC (mean = 22.1), followed by ABC (mean = 21.7), EFV (mean = 10.4), and LPV/r (mean = 4.00). Several subjects, particularly subject 92, had markedly high levels of archived drug resistance (Table 3).
Proviral diversity and divergence do not correlate with demographic immune characteristics.
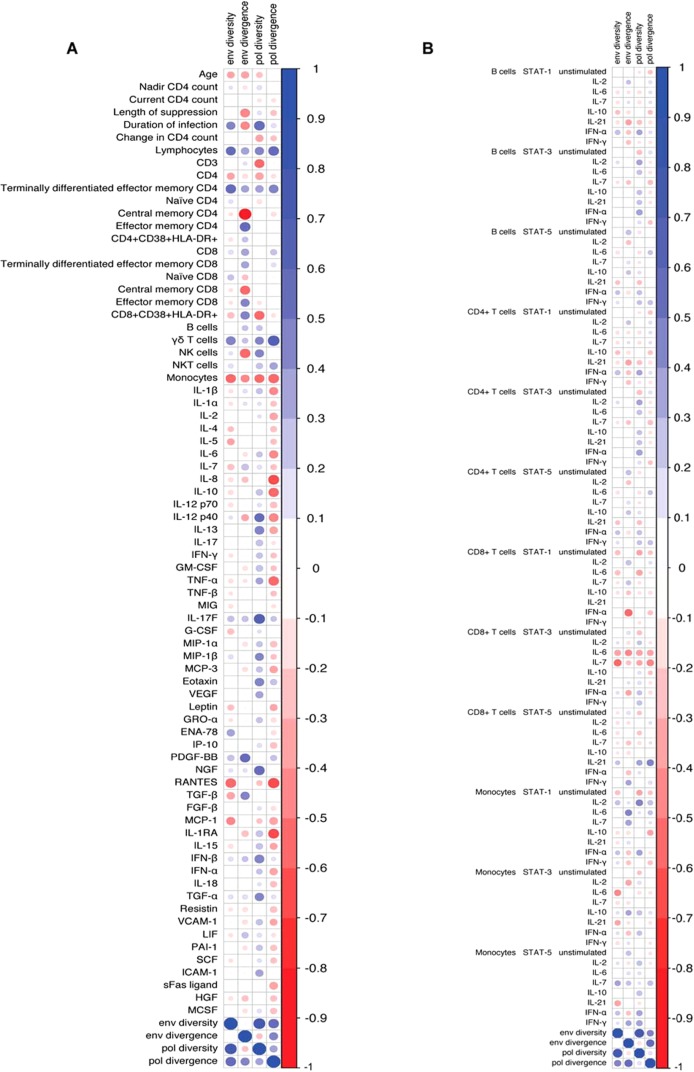
Demographic data, immune cell phenotyping, serum cytokine levels, and phospho-flow analysis of intracellular signaling responses were used to determine whether these characteristics associate with proviral diversity and divergence in env and pol (Fig. 3; see also Fig. S1 to S5 in the supplemental material). The diversity and divergence in env and pol were positively correlated (Fig. 3). In addition, there were positive correlations between the percentage of total lymphocytes and the percentage of CD4+ effector memory cells with proviral diversity and divergence. The percentage of monocytes was inversely correlated with proviral diversity and divergence. Proviral diversity and divergence were also weakly associated with decreased levels of CD8+ T cell STAT-3 responses to IL-6 and IL-7. After control of the false-discovery rate to be no more than 10%, none of the associations reached statistical significance. In addition, none of the demographic characteristics such as age, nadir CD4+ T cell count, or duration of infection were significantly associated with the proviral diversity or divergence in env or pol. Thus, overall there were no statistically significant correlations between proviral diversity and divergence and any demographic or immune characteristic (Fig. 3). In addition, neither ART resistance levels nor coreceptor tropism correlated with any of the immunological or demographic parameters (data not shown).
FIG 3.
Correlation matrices of proviral characteristics and immune characteristics. The colored circles represent the Spearman's correlation coefficient, sized according the magnitude of the coefficient and colored according to the scale bar shown on the right. Positive correlations are blue and negative are red, and the size denotes the magnitude of the correlation. (A) Correlation matrix of proviral characteristics and cellular markers and plasma cytokine levels; (B) correlation matrix of proviral characteristics and signaling markers.
Virologic failure does not predict level of immune activation.
As the characteristics of the virus were not significantly correlated with the level of ongoing immune activation in this cohort of well-controlled, HIV-infected individuals, we examined whether a clinical history of virologic failure was associated with immune activation. We performed Wilcoxon rank sum tests comparing the proviral and immune characteristics between subjects without and with VF while controlling the false-discovery rate to be no more than 10% (Table 4). A history of virologic failure was associated with a trend for increased pol diversity, increased drug resistance, and duration of infection, indicating that our data set was adequately robust to detect some viral characteristics associated with clinical history. However, for the remaining proviral and immune characteristics, individuals with a history of virologic failure were not significantly different from individuals without a history of virologic failure (Table 4 and data not shown). This result held even for immune characteristics that have previously been shown to be markers of HIV-associated immune activation, such as the percentage of CD38- and HLA-DR-expressing CD4+ and CD8+ T cells. Thus, a clinical history of virologic failure does not predict the level of immune activation in our cohort.
TABLE 4.
Comparison of viral and immune characteristics between subjects without and with a history of virologic failure
| Characteristic | No VF |
VF |
Wilcoxon P value | Adjusted P valuea | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| % CD3+ | 67.0 | 12.3 | 72.7 | 6.44 | 0.64 | 0.78 |
| % CD4+ | 30.2 | 9.36 | 26.8 | 12.51 | 0.54 | 0.78 |
| % CD8+ | 35.7 | 10.6 | 42.6 | 12.84 | 0.46 | 0.78 |
| % HLA-DR+ CD38+ CD4+ | 19.5 | 10.6 | 16.2 | 6.14 | 0.78 | 0.78 |
| % HLA-DR+ CD38+ CD8+ | 23.5 | 11.0 | 17.5 | 6.31 | 0.15 | 0.34 |
| Plasma IL-6 level (pg/ml) | 0.5 | 0.22 | 0.49 | 0.25 | 0.68 | 0.78 |
| Baseline CD4+ STAT-3 activity | 0.750 | 0.53 | 0.76 | 0.5 | 0.78 | 0.78 |
| CD4+ STAT-3 activity in response to IL-6 | −0.810 | 0.43 | −0.82 | 0.44 | 0.78 | 0.78 |
| env diversity (%) | 2.57 | 1.62 | 3.66 | 1.72 | 0.12 | 0.31 |
| env divergence (%) | 10.1 | 0.60 | 9.32 | 0.96 | 0.28 | 0.56 |
| pol diversity (%) | 1.03 | 0.39 | 1.69 | 0.63 | 0.09 | 0.28 |
| pol divergence (%) | 3.97 | 0.29 | 4.22 | 0.61 | 0.69 | 0.78 |
| LPV/r resistance score | 0.140 | 0.38 | 7.37 | 20.3 | 0.56 | 0.78 |
| 3TC resistance score | 1.12 | 1.45 | 40.5 | 28.4 | 0.008 | 0.09 |
| ABC resistance score | 4.43 | 6.31 | 36.7 | 26.8 | 0.01 | 0.09 |
| EFV resistance score | 0.690 | 1.29 | 19.0 | 26.8 | 0.08 | 0.28 |
| Length of suppression (mo) | 31 | 44.5 | 46 | 25.7 | 0.01 | 0.09 |
| Duration of infection (yr) | 11.6 | 5.01 | 18 | 4.6 | 0.04 | 0.17 |
Adjusted P values account for a false-discovery rate of 10%.
DISCUSSION
Recently, much HIV-1 research has focused on defining the mechanisms driving HIV-associated immune activation, which is associated with early manifestations of aging, such as increased risk of developing CVD. We report here an evaluation of the relationship between proviral characteristics and markers of immune activation in a cohort of virologically suppressed HIV-infected individuals on ART. Our study is the first to investigate HIV-associated immune activation in the context of proviral populations in effectively treated HIV-infected individuals and is the most comprehensive evaluation of immune function during chronic HIV-1 infection to date. As some studies have suggested that cryptic viral replication in virologically suppressed individuals may occur and contributes to chronic immune activation (44), and even fully virologically suppressed individuals have increased levels of soluble activation markers (58), we hypothesized that by examining the relationship between the proviral population and immune activation we could reveal previously undiscovered contributors to HIV-associated immune activation. We found that subjects, whether or not they had a history of virologic failure, exhibited a wide range of diversity and divergence in both env and pol. We also found that proviral diversity and divergence in env and pol, coreceptor tropism, level of drug resistance, and a history of virologic failure do not correlate with markers of immune activation. These results indicate that the nature of the latent virus does not appear to be a major driver of HIV-associated immune activation and its effect on the health of HIV-infected individuals, suggesting that other factors drive the immune activation that persists despite effective treatment.
This evaluation of proviral diversity and divergence in the setting of effective ART provides new insight into the nature of the archived virus during long-term, fully suppressive treatment. Relatively few studies have evaluated such characteristics during treatment, particularly with modern regimens. Studies performed by Finzi et al. in 1997 and 1999 indicated that latent provirus represents a relatively stable population containing archived viruses from prior to initiation of therapy (47, 48). Subsequent studies have revealed that the proviral HIV-1 DNA sequences can remain dynamic even during antiretroviral therapy, acting as an archive of circulating plasma viruses found throughout the course of HIV infection (49, 50). However, given that HIV-1 diversity accumulates more quickly in acute infection (59), it is likely that much of the env and pol diversity and divergence observed in our cohort accumulated prior to ART initiation. We were unable to directly assess this, as we did not have a reliable history documenting the duration of HIV-1 infection prior to ART initiation.
Within this framework, the subjects in our study exhibited a wide range of both diversity and divergence in their proviral populations during effective treatment. The diversity we observed in both the env and pol genes was similar to the levels of proviral diversity previously reported during antiretroviral therapy (49, 60) but higher than the levels observed when treatment is started during acute infection (61). Thus, our data support the possibility that the proviral population may be very diverse and divergent even if the individual has been effectively treated with modern ART regimens. The lack of association between these viral characteristics and immune activation in our study suggests that even effective viral suppression may not affect already established immunological changes originating from HIV-1 infection.
Several prior studies suggest that factors other than proviral diversity and divergence, including coreceptor tropism and the presence of drug resistance mutations, are associated with immune activation. For instance, two studies suggested that coreceptor tropism in untreated, HIV-infected adults was associated with immune activation (34, 62). However, in both our study and a recent study by Saracino et al. (35), the presence of CXCR4-tropic viruses in proviral DNA was not associated with the level of immune activation in individuals with fully suppressed or very low viral loads. Thus, the association between CXCR4 tropism and inflammation appears to require active viral replication. Additionally, in viremic individuals, the presence of drug resistance mutations has been associated with lower levels of immune activation, rates of CD4+ T cell depletion, and progression to AIDS, independently of HIV RNA levels (28–31). Our study was the first to examine archived drug resistance mutations in individuals on fully suppressive treatment, and we did not identify a significant association between drug resistance mutations and inflammation.
Interestingly, we also found that a clinical history of virologic failure was not significantly associated with increased levels of immune activation, suggesting that immunological changes established early during infection or in the pretreatment phase may set the stage for ongoing inflammation. Such changes appear to be unrelated to the viral characteristics and inadequate to control the infection regardless of magnitude. This finding is consistent with studies of blood and gut-associated lymphoid tissue sequences, in which no significant changes in diversity were observed after treatment initiation, and immune activation persisted despite ART (63).
The limitations of our study include our small sample size and our lack of longitudinal data. Assuming two normally distributed random variables, with our sample size of 16, we would have 88.2% power to detect a strong correlation, 0.7 or larger, but only 50.5% power to detect the more modest correlation of 0.5. We did find trends and/or statistically significant differences between subjects with and without a history of VF in pol diversity, ART resistance scores, and duration of infection. An increased sample size or longitudinal data may have nonetheless increased our ability to find significant associations between the nature of the provirus and immune activation and/or associations between the nature of the provirus and changes in immune activation over time. In addition, due to limitations in sample volume, we had a relatively modest number of HIV-1 sequences with which to calculate diversity and divergence for some individuals. However, exclusion of subjects with low numbers of sequences did not alter results (data not shown). Finally, viral characteristics that we did not measure, such as escape mutations from the immune response within each individual, could be the primary driving force behind immune activation. It is also important to note that researchers have yet to establish a common, widely accepted definition of immune activation that effectively encompasses molecular and cellular characteristics. Studies of HIV-associated immune activation have focused on different aspects of the immune system such as expression of activation markers on immune cells, cytokine levels, and functional responses. While numerous biomarkers, such as IL-6, soluble CD14, and HLA-DR+ CD38+ CD4+ and CD8+ T cells, predict disease progression, we have yet to discover how they interact with each other (1). A key strength of our study is that it is the most comprehensive evaluation of immune status and function performed to date with treated HIV-infected individuals, as it accounts for cellular subsets, signaling pathways, and cytokine profiles, while previous studies have focused on only one or two of these features at a time.
By showing that characteristics of the proviral population, including viral diversity, divergence, coreceptor tropism, and the frequency of drug resistance mutations, do not significantly correlate with levels of immune activation in this modestly sized cohort of ART-treated HIV-1-infected individuals, our study reveals that these viral characteristics may not be the most significant drivers of chronic immune activation. Furthermore, a clinical history of virologic failure does not permanently imprint the immune system through increased levels of immune activation if the virus is currently suppressed. Our study therefore has important implications for treatment decisions and outcomes, indicating that a history of virologic failure or high viral diversity is not inexorably linked to increased immune activation as long as appropriate suppressive ART regimens are provided. Future studies should extend these findings to larger longitudinal cohorts followed prior to and after successful viral suppression with ART.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Translational Medicine and Applied Research grant from the Stanford University School of Medicine, Department of Medicine, to C.A.B., A.R.Z., and D.A.K. and NIH U19 grants AI057240 and AI090019 to M.M.D. E.C.L. was supported by an undergraduate Major Grant from Stanford University, L.S. by the Stanford University School of Medicine MedScholars Program, N.L.B. by a Stanford Graduate Fellowship, D.M.S.-A. by NSF training grant DGE-114740, and D.F. by a fellowship from the Stanford Center on Longevity. The Stanford HIV Aging Cohort was supported by California HIV Research Program grant ID10-ST-025 and National Institutes of Health supplement AI069556.
We thank the clinical and laboratory staff, especially Maya Balamane and Meg Winslow for sample processing and storage, and Robert Shafer for useful advice in scoring drug resistance. We gratefully acknowledge the individuals who participated in our study.
None of us has a conflict of interest in this study.
Footnotes
Published ahead of print 21 May 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01257-14.
REFERENCES
- 1.Paiardini M, Müller-Trutwin M. 2013. HIV-associated chronic immune activation. Immunol. Rev. 254:78–101. 10.1111/imr.12079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.d'Ettorre G, Paiardini M, Ceccarelli G, Silvestri G, Vullo V. 2011. HIV-associated immune activation: from bench to bedside. AIDS Res. Hum. Retroviruses 27:355–364. 10.1089/aid.2010.0342 [DOI] [PubMed] [Google Scholar]
- 3.2013. Immune responses and cell signaling during chronic HIV infection. InTech 10.5772/53010 [DOI] [Google Scholar]
- 4.Lawn SD, Butera ST, Folks TM. 2001. Contribution of immune activation to the pathogenesis and transmission of human immunodeficiency virus type 1 infection. Clin. Microbiol. Rev. 14:753–777. 10.1128/CMR.14.4.753-777.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sodora DL, Silvestri G. 2008. Immune activation and AIDS pathogenesis. AIDS 22:439–446. 10.1097/QAD.0b013e3282f2dbe7 [DOI] [PubMed] [Google Scholar]
- 6.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, Mempel TR. 2012. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 490:283–289. 10.1038/nature11398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, JAcobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, Wolinsky SM. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 179:859–869. 10.1086/314660 [DOI] [PubMed] [Google Scholar]
- 8.Leng Q, Borkow G, Weisman Z, Stein M, Kalinkovich A, Bentwich Z. 2001. Immune activation correlates better than HIV plasma viral load with CD4 T-cell decline during HIV infection. J. Acquir. Immune Defic. Syndr. 27:389–397. 10.1097/00126334-200108010-00010 [DOI] [PubMed] [Google Scholar]
- 9.Deeks SG, Kitchen CMR, Liu L, Guo H, Gascon R, Narváez AB, Hunt P, Martin JN, Kahn JO, Levy J, McGrath MS, Hecht FM. 2004. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood 104:942–947. 10.1182/blood-2003-09-3333 [DOI] [PubMed] [Google Scholar]
- 10.Kuller LH, Tracy R, Belloso W, De Wit S, Drummond F, Lane HC, Ledergerber B, Lundgren J, Neuhaus J, Nixon D, Paton NI, Neaton JD, INSIGHT SMART Study Group 2008. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 5:e203. 10.1371/journal.pmed.0050203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stein JH, Hsue PY. 2012. Inflammation, immune activation, and CVD risk in individuals with HIV infection. JAMA 308:405–406. 10.1001/jama.2012.8488 [DOI] [PubMed] [Google Scholar]
- 12.Subramanian S, Tawakol A, Burdo TH, Abbara S, Wei J, Vijayakumar J, Corsini E, Abdelbaky A, Zanni MV, Hoffmann U, Williams KC, Lo J, Grinspoon SK. 2012. Arterial inflammation in patients with HIV. JAMA 308:379–386. 10.1001/jama.2012.6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tebas P, Henry WK, Matining R, Weng-Cherng D, Schmitz J, Valdez H, Jahed N, Myers L, Powderly WG, Katzenstein D. 2008. metabolic and immune activation effects of treatment interruption in chronic HIV-1 infection: implications for cardiovascular risk. PLoS One 3:e2021. 10.1371/journal.pone.0002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fahey JL, Taylor JMG, Manna B, Nishanian P, Aziz N, Giorgi JV, Detels R. 1998. Prognostic significance of plasma markers of immune activation, HIV viral load and CD4 T-cell measurements. AIDS 12:1581–1590. 10.1097/00002030-199813000-00004 [DOI] [PubMed] [Google Scholar]
- 15.Triant VA. 2013. Cardiovascular disease and HIV infection. Curr. HIV/AIDS Rep. 10:199–206. 10.1007/s11904-013-0168-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Triant VA, Lee H, Hadigan C, Grinspoon SK. 2007. Increased acute myocardial infarction rates and cardiovascular risk factors among patients with human immunodeficiency virus disease. J. Clin. Endocrinol. Metab. 92:2506–2512. 10.1210/jc.2006-2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fichtenbaum CJ. 2011. Inflammatory markers associated with coronary heart disease in persons with HIV infection. Curr. Infect. Dis. Rep. 13:94–101. 10.1007/s11908-010-0153-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright EJ, Grund B, Robertson K, Brew BJ, Roediger M, Bain MP, Drummond F, Vjecha MJ, Hoy J, Miller C, Penalva de Oliveira AC, Pumpradit W, Shlay JC, El-Sadr W, Price RW, INSIGHT SMART Study Group 2010. Cardiovascular risk factors associated with lower baseline cognitive performance in HIV-positive persons. Neurology 75:864–873. 10.1212/WNL.0b013e3181f11bd8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulliam L, Gascon R, Stubblebine M, McGuire D, McGrath MS. 1997. Unique monocyte subset in patients with AIDS dementia. Lancet 349:692–695. 10.1016/S0140-6736(96)10178-1 [DOI] [PubMed] [Google Scholar]
- 20.Gazzola L, Bellistri GM, Tincati C, Ierardi V, Savoldi A, del Dole A, Tagliabue L, d'Arminio Monforte A, Marchetti G. 2013. Association between peripheral T-lymphocyte activation and impaired bone mineral density in HIV-infected patients. J. Transl. Med. 11:51. 10.1186/1479-5876-11-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desquilbet L, Margolick JB, Fried LP, Phair JP, Jamieson BD, Holloway M, Jacobson LP, Multicenter AIDS Cohort Study. 2009. Relationship between a frailty-related phenotype and progressive deterioration of the immune system in HIV-infected men. J. Acquir. Immune Defic. Syndr. 50:299–306. 10.1097/QAI.0b013e3181945eb0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erlandson KM, Allshouse AA, Jankowski CM, Lee EJ, Rufner KM, Palmer BE, Wilson CC, MaWhinney S, Kohrt WM, Campbell TB. 2013. Association of functional impairment with inflammation and immune activation in HIV type 1-infected adults receiving effective antiretroviral therapy. J. Infect. Dis. 208:249–259. 10.1093/infdis/jit147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagar M, Lavreys L, Baeten JM, Richardson BA, Mandaliya K, Chohan BH, Kreiss JK, Overbaugh J. 2003. Infection with multiple human immunodeficiency virus type 1 variants is associated with faster disease progression. J. Virol. 77:12921–12926. 10.1128/JVI.77.23.12921-12926.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piantadosi A, Chohan B, Panteleeff D, Baeten JM, Mandaliya K, Ndinya-Achola JO, Overbaugh J. 2009. HIV-1 evolution in gag and env is highly correlated but exhibits different relationships with viral load and the immune response. AIDS 23:579–587. 10.1097/QAD.0b013e328328f76e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalmet K, Dauwe K, Foquet L, Baatz F, Seguin-Devaux C, Van Der Gucht B, Vogelaers D, Vandekerckhove L, Plum J, Verhofstede C. 2012. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J. Infect. Dis. 205:174–184. 10.1093/infdis/jir714 [DOI] [PubMed] [Google Scholar]
- 26.Richman DD, Bozzette SA. 1994. The impact of the syncytium-inducing phenotype of human immunodeficiency virus on disease progression. J. Infect. Dis. 169:968–974. 10.1093/infdis/169.5.968 [DOI] [PubMed] [Google Scholar]
- 27.D'Aquila RT, Johnson VA, Welles SL, Japour AJ, Kuritzkes DR, DeGruttola V, Reichelderfer PS, Coombs RW, Crumpacker CS, Kahn JO, Richman DD. 1995. Zidovudine resistance and HIV-1 disease progression during antiretroviral therapy. Ann. Intern. Med. 122:401–408. 10.7326/0003-4819-122-6-199503150-00001 [DOI] [PubMed] [Google Scholar]
- 28.Wang R, Bosch RJ, Benson CA, Lederman MM. 2012. Drug-resistant virus has reduced ability to induce immune activation. J. Acquir. Immune Defic. Syndr. 61(4):e60–e63. 10.1097/QAI.0b013e31827171d7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt PW, Deeks SG, Bangsberg DR, Moss A, Sinclair E, Liegler T, Bates M, Tsao G, Lampiris H, Hoh R, Martin JN. 2006. The independent effect of drug resistance on T cell activation in HIV infection. AIDS 20:691–699. 10.1097/01.aids.0000216369.30948.18 [DOI] [PubMed] [Google Scholar]
- 30.Grabar S. 2000. Clinical outcome of patients with HIV-1 infection according to immunologic and virologic response after 6 months of highly active antiretroviral therapy. Ann. Intern. Med. 133:401–410. 10.7326/0003-4819-133-6-200009190-00007 [DOI] [PubMed] [Google Scholar]
- 31.Miller V, Phillips AN, Clotet B, Mocroft A, Ledergerber B, Kirk O, Ormaasen V, Gargalianos-Kakolyris P, Vella S, Lundgren JD. 2002. Association of virus load, CD4 cell count, and treatment with clinical progression in human immunodeficiency virus-infected patients with very low CD4 cell counts. J. Infect. Dis. 186:189–197. 10.1086/341466 [DOI] [PubMed] [Google Scholar]
- 32.Lucas GM, Gallant JE, Moore RD. 2004. Relationship between drug resistance and HIV-1 disease progression or death in patients undergoing resistance testing. AIDS 18:1539. 10.1097/01.aids.0000131339.68666.1a [DOI] [PubMed] [Google Scholar]
- 33.Campbell TB, Shulman NS, Johnson SC, Zolopa AR, Young RK, Bushman L, Fletcher CV, Lanier ER, Merigan TC, Kuritzkes DR. 2005. Antiviral activity of lamivudine in salvage therapy for multidrug-resistant HIV-1 infection. Clin. Infect. Dis. 41:236–242. 10.1086/430709 [DOI] [PubMed] [Google Scholar]
- 34.Rozera G, Abbate I, Vlassi C, Giombini E, Lionetti R, Selleri M, Zaccaro P, Bartolini B, Corpolongo A, D'Offizi G, Baiocchini A, Del Nonno F, Ippolito G, Capobianchi MR. 2014. Quasispecies tropism and compartmentalization in gut and peripheral blood during early and chronic phases of HIV-1 infection: possible correlation with immune activation markers. Clin. Microbiol. Infect. 20:O157–O166. 10.1111/1469-0691.12367 [DOI] [PubMed] [Google Scholar]
- 35.Saracino A, Monno L, Scudeller L, Bruno G, Ladisa N, Punzi G, Volpe A, Lagioia A, Angarano G. 2013. X4 viruses are frequently archived in patients with long-term HIV infection but do not seem to influence the “inflamm-aging” process. BMC Infect. Dis. 13:220. 10.1186/1471-2334-13-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yehia BR, Fleishman JA, Metlay JP, Moore RD, Gebo KA. 2012. Sustained viral suppression in HIV-infected patients receiving antiretroviral therapy. JAMA 308:339–342. 10.1001/jama.2012.5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chun TW. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Invest. 115:3250–3255. 10.1172/JCI26197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Günthard HF, Frost SD, Leigh-Brown AJ, Ignacio CC, Kee K, Perelson AS, Spina CA, Havlir DV, Hezareh M, Looney DJ, Richman DD, Wong JK. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J. Virol. 73:9404–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. 1998. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 95:8869–8873. 10.1073/pnas.95.15.8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chun T-W, Justement JS, Moir S, Hallahan CW, Maenza J, Mullins JI, Collier AC, Corey L, Fauci AS. 2007. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195:1762–1764. 10.1086/518250 [DOI] [PubMed] [Google Scholar]
- 41.Martínez MA, Cabana M, Ibáñez A, Clotet B, Arnó A, Ruiz L. 1999. Human immunodeficiency virus type 1 genetic evolution in patients with prolonged suppression of plasma viremia. Virology 256:180–187. 10.1006/viro.1999.9601 [DOI] [PubMed] [Google Scholar]
- 42.Sharkey M, Triques K, Kuritzkes DR, Stevenson M. 2005. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J. Virol. 79:5203–5210. 10.1128/JVI.79.8.5203-5210.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. 2008. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 105:3879–3884. 10.1073/pnas.0800050105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buzón MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, Palmer S, Stevenson M, Clotet B, Blanco J, Martinez-Picado J. 2010. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat. Med. 16:460–465. 10.1038/nm.2111 [DOI] [PubMed] [Google Scholar]
- 45.Mavigner M, Delobel P, Cazabat M, Dubois M, L'Faqihi-Olive F-E, Raymond S, Pasquier C, Marchou B, Massip P, Izopet J. 2009. HIV-1 residual viremia correlates with persistent T-cell activation in poor immunological responders to combination antiretroviral therapy. PLoS One 4:e7658. 10.1371/journal.pone.0007658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saison J, Ferry T, Demaret J, Maucort Boulch D, Venet F, Perpoint T, Ader F, Icard V, Chidiac C, Monneret G, Lyon HIV Cohort Study. 2014. Association between discordant immunological response to HAART, regulatory T cells percentage, immune cell activation and very low level viremia in HIV-infected patients. Clin. Exp. Immunol. 176:401–409. 10.1111/cei.12278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick JB, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295 [DOI] [PubMed] [Google Scholar]
- 48.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517. 10.1038/8394 [DOI] [PubMed] [Google Scholar]
- 49.Lambotte O, Chaix M-L, Gubler B, Nasreddine N, Wallon C, Goujard C, Rouzioux C, Taoufik Y, Delfraissy J-F. 2004. The lymphocyte HIV reservoir in patients on long-term HAART is a memory of virus evolution. AIDS 18:1147–1158. 10.1097/00002030-200405210-00008 [DOI] [PubMed] [Google Scholar]
- 50.Buzón MJ, Codoñer FM, Frost SDW, Pou C, Puertas MC, Massanella M, Dalmau J, Llibre JM, Stevenson M, Blanco J, Clotet B, Paredes R, Martinez-Picado J. 2011. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog. 7:e1002314. 10.1371/journal.ppat.1002314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, Persaud D, Gallant JE, Cofrancesco J, Quinn TC, Wilke CO, Ray SC, Siliciano JD, Nettles RE, Siliciano RF. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441–6457. 10.1128/JVI.00591-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parera M, Ibáñez A, Clotet B, Martinez MA. 2004. Lack of evidence for protease evolution in HIV-1-infected patients after 2 years of successful highly active antiretroviral therapy. J. Infect. Dis. 189:1444–1451. 10.1086/382485 [DOI] [PubMed] [Google Scholar]
- 53.Joos B, Fischer M, Kuster H, Pillai SK, Wong JK, Böni J, Hirschel B, Weber R, Trkola A, Gunthard HF, Swiss HIV Cohort Study. 2008. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci. U. S. A. 105:16725–16730. 10.1073/pnas.0804192105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kieffer TL, Finucane MM, Nettles RE, Quinn TC, Broman KW, Ray SC, Persaud D, Siliciano RF. 2004. Genotypic analysis of HIV-1 drug resistance at the limit of detection: virus production without evolution in treated adults with undetectable HIV loads. J. Infect. Dis. 189:1452–1465. 10.1086/382488 [DOI] [PubMed] [Google Scholar]
- 55.Sedaghat AR, Siliciano JD, Brennan TP, Wilke CO, Siliciano RF. 2007. Limits on replenishment of the resting CD4+ T cell reservoir for HIV in patients on HAART. PLoS Pathog. 3:e122. 10.1371/journal.ppat.0030122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krutzik PO, Nolan GP. 2006. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat. Methods 3:361–368. 10.1038/nmeth872 [DOI] [PubMed] [Google Scholar]
- 57.Furman D, Jojic V, Kidd B, Shen-Orr S, Price J, Jarrell J, Tse T, Huang H, Lund P, Maecker HT, Utz PJ, Dekker CL, Koller D, Davis MM. 2013. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol. Syst. Biol. 9:659. 10.1038/msb.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ostrowski SR, Katzenstein TL, Pedersen BK, Gerstoft J, Ullum H. 2008. Residual viraemia in HIV-1-infected patients with plasma viral load ≤20 copies/ml is associated with increased blood levels of soluble immune activation markers. Scand. J. Immunol. 68:652–660. 10.1111/j.1365-3083.2008.02184.x [DOI] [PubMed] [Google Scholar]
- 59.Maldarelli F, Kearney M, Palmer S, Stephens R, Mican J, Polis MA, Davey RT, Kovacs J, Shao W, Rock-Kress D, Metcalf JA, Rehm C, Greer SE, Lucey DL, Danley K, Alter H, Mellors JW, Coffin JM. 2013. HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. J. Virol. 87:10313–10323. 10.1128/JVI.01225-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ibáñez A, Clotet B, Martínez MA. 2001. Absence of genetic diversity reduction in the HIV-1 integrated proviral LTR sequence population during successful combination therapy. Virology 282:1–5. 10.1006/viro.2000.0840 [DOI] [PubMed] [Google Scholar]
- 61.Gall A, Kaye S, Hué S, Bonsall D, Rance R, Baillie GJ, Fidler SJ, Weber JN, McClure MO, Kellam P, SPARTAC Trial Investigators 2013. Restriction of V3 region sequence divergence in the HIV-1 envelope gene during antiretroviral treatment in a cohort of recent seroconverters. Retrovirology 10:8. 10.1186/1742-4690-10-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Portales P, Psomas KC, Tuaillon E, Mura T, Vendrell J-P, Eliaou J-F, Reynes J, Corbeau P. 2012. The intensity of immune activation is linked to the level of CCR5 expression in human immunodeficiency virus type 1-infected persons. Immunology 137:89–97. 10.1111/j.1365-2567.2012.03609.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, Mohri H, Markowitz M. 2012. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 8:e1002506. 10.1371/journal.ppat.1002506 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.