FIG 5.
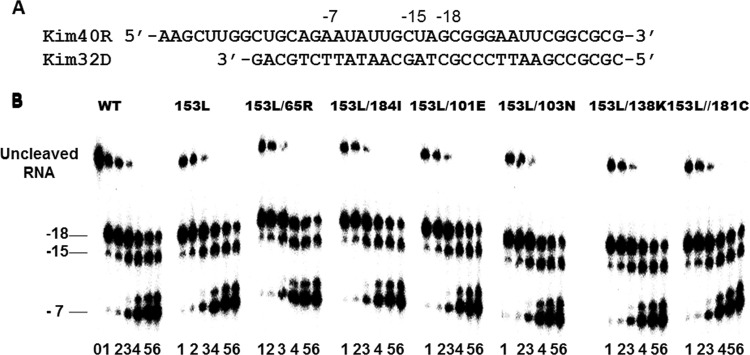
RNase H activities of WT and mutant recombinant RT enzymes. (A) Graphic representation of the RNA/DNA (kim40R/kim32D) substrate duplex used to monitor the cleavage efficiencies of mutant and WT RTs. Positions of cleavage sites relative to the 3′ end of the primer are shown at the top. The 40-mer RNA kim40R was labeled at its 5′ terminus by 32P and annealed to a 32-mer DNA oligonucleotide (kim32D). (B) RNase H activity was analyzed by monitoring substrate cleavage at 0.2, 0.5, 1, 3, 5, and 10 min (lanes 1 to 6, respectively) in time course experiments. Lane 0 (time point 0) shows the uncleaved substrate in a control reaction without RT enzyme. The uncleaved substrate and cleaved products relative to the 3′ terminus of the DNA primer are indicated on the left. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis. Experiments were repeated at least twice with similar results being obtained each time. The gel is from a representative experiment.
