LETTER
Fosfomycin (FOM) is an antibiotic produced by Streptomyces fradiae (1) and was approved for clinical use in Japan in 1980. FOM blocks MurA, which mediates bacterial peptidoglycan biosynthesis in its early step, showing a broad-spectrum antimicrobial activity against both Gram-positive and Gram-negative bacteria. FOM penetrates into bacterial cells via sugar transporters, such as GlpT and UhpT, located at the cytoplasmic membrane, and spontaneous FOM-resistant mutants appear due to a reduction or lack of these transporters. Moreover, several enzymes, such as FosA, FosB, FosC, FosD, FomX, FomA, and FomB, have been reported, and FosA was first characterized as a glutathione S-transferase of FOM (2) (Fig. 1). After our first report about FosA3 and FosC2 in 2010 (3), FosA3-producing Escherichia coli isolates were recovered from humans, livestock, and/or pets (4–7), and the fosA3 gene has already transferred to Klebsiella pneumoniae (6) by a probable IS26 composite transposon carrying fosA3.
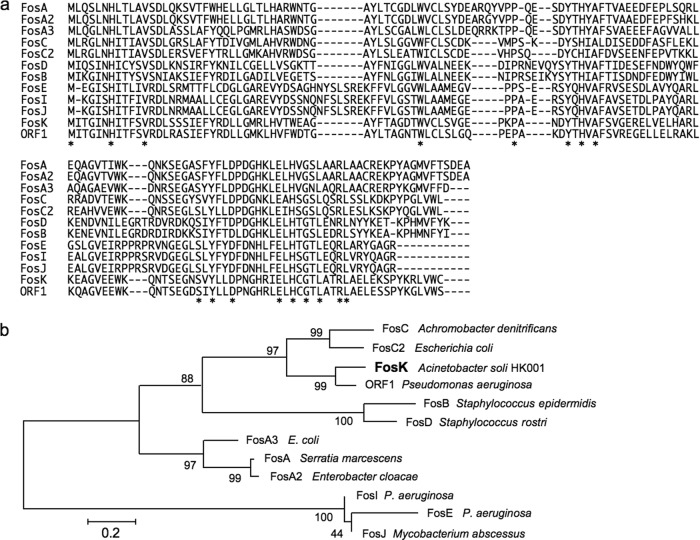
FIG 1.
(a) Predicted amino acid sequences of FosK and other fosfomycin-modifying enzymes. *, amino acid residue conserved among the 12 fosfomycin resistance determinants. (b) Phylogenic relationships among the 12 glutathione S-transferases, including probable ones calculated by MEGA 5 (http://www.megasoftware.net/). GenBank or Protein Data Bank accession numbers are indicated for the following proteins: FosA (AAA98399), FosA2 (ACC85616), FosA3 (AB522970), FosB (CAA38136), FosC (AAZ14834), FosC2 (AB522969), FosD (AHB87392), FosE (BAO48025), FosI (BAO47999), FosJ (YP_006316014), FosK (AB917040), and ORF1 (AAP50248).
Acinetobacter soli HK001 was isolated from a blood culture of an infected human, and it showed very high resistance to FOM (MIC, >8,000 μg/ml) according to the agar dilution method recommended by the CLSI (8) in the presence of glucose-6-phosphate (G6P) (25 μg/ml), which induces UhpT. Four amplicons of class 1 integrons were found by PCR using 2 primers, 5′CS-Class1-integron (5′-GGCATCCAAGCAGCAAG-3′) and 3′CS-Class1-integron (5′-AAGCAGACTTGACCTGA-3′). An amplicon of 1.2 kb was excised and purified. Its nucleotide sequence was directly determined and revealed an aacA4 gene and a new gene cassette located between intI1 and the 3′-CS (conserved sequence). The new cassette encoded a protein with significant similarity to other Fos proteins (Fig. 1) and was named FosK. The deduced amino acid sequence of FosK showed 81% identity in its amino acid sequence to open reading frame 1 (ORF1) of Pseudomonas aeruginosa (9). Moreover, 52%, 52%, 51%, 50%, 48%, and 47% amino acid identities were observed between FosK and FosC2, FosD, FosA3, FosA, FosA2, and FosC, respectively, suggesting their close phylogenic relationship (Fig. 1). The fosK gene was again amplified by PCR using total bacterial DNA and a high-fidelity DNA polymerase, PrimeSTAR HS (TaKaRa Bio Inc., Ohtsu, Japan), together with primers F2-BamHI (5′-CGGGATCCCGACATGGTTCAAACACGCCAGGC-3′) and R2-HindIII (5′-TACCCAAGCTTGGGTTTTGGGGCGGACTTGTAGC-3′). The amplicon was ligated with pBCSK+ and cleaved by BamHI and HindIII, and E. coli DH10B was transformed with the recombinant plasmids. Then FOM-resistant transformants were selected. After nucleotide sequencing of the insert on both strands, a clone carrying no mutation in the fosK gene was finally chosen. The FOM MIC for the transformant harboring intact fosK was augmented to >2,048 μg/ml from 1 μg/ml for the recipient with G6P (25 μg/ml) (Table 1).
TABLE 1.
MICs of fosfomycin for A. soli HK001 and E. coli DH10B transformed with the fosK gene
| Strain | FOM MIC (μg/ml)a |
|---|---|
| Acinetobacter soli HK001 | >8,192 |
| E. coli DH10B | 1 |
| E. coli DH10B(pBCSK+) | 2 |
| E. coli DH10B(pBCSK+::fosK) | >2,048 |
| E. coli ATCC 25922 | 2 |
FOM, fosfomycin. MICs were measured by the agar dilution method recommended by the CLSI.
FOM was recently considered to be a potent agent for treatment of infections caused by multidrug-resistant bacteria, such as extended-spectrum β-lactamase (ESBL)-producing E. coli and K. pneumoniae (10). FOM has also been approved for veterinary use in various countries (11). The fosK gene, together with aacA4, is mediated by a class 1 integron, and thus this genetic element will be further transmitted into various Enterobacteriaceae. Since fosK confers on bacteria a very high level of resistance to fosfomycin, we should diligently monitor the prevalence and trend of fosK as well as of fosA3 in both human and animals going forward.
Nucleotide sequence accession number.
The fosK gene has been assigned accession number AB917040.
ACKNOWLEDGMENT
This study was supported by grant no. H24-Shinko-Ippan-010.
Footnotes
Published ahead of print 19 May 2014
REFERENCES
- 1.Rogers TO, Birnbaum J. 1974. Biosynthesis of fosfomycin by Streptomyces fradiae. Antimicrob. Agents Chemother. 5:121–132. 10.1128/AAC.5.2.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernat BA, Laughlin LT, Armstrong RN. 1997. Fosfomycin resistance protein (FosA) is a manganese metalloglutathione transferase related to glyoxalase I and the extradiol dioxygenases. Biochemistry 36:3050–3055. 10.1021/bi963172a [DOI] [PubMed] [Google Scholar]
- 3.Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. 2010. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob. Agents Chemother. 54:3061–3064. 10.1128/AAC.01834-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou J, Yang X, Zeng Z, Lv L, Yang T, Lin D, Liu JH. 2013. Detection of the plasmid-encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J. Antimicrob. Chemother. 68:766–770. 10.1093/jac/dks465 [DOI] [PubMed] [Google Scholar]
- 5.Pan YS, Yuan L, Zong ZY, Liu JH, Wang LF, Hu GZ. 2014. A multidrug-resistance region containing blaCTX-M-65, fosA3 and rmtB on conjugative IncFII plasmids in Escherichia coli ST117 isolates from chicken. J. Med. Microbiol. 63:485–488. 10.1099/jmm.0.070664-0 [DOI] [PubMed] [Google Scholar]
- 6.Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, Arakawa Y. 2012. Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67:2843–2847. 10.1093/jac/dks319 [DOI] [PubMed] [Google Scholar]
- 7.Sato N, Kawamura K, Nakane K, Wachino J, Arakawa Y. 2013. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resist. 19:477–482. 10.1089/mdr.2013.0061 [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility testing of bacteria that grow aerobically. Approved standard, 8th ed. Document M7-A9 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9.Yatsuyanagi J, Saito S, Konno T, Harata S, Suzuki N, Amano K. 2005. The ORF1 gene located on the class-1-integron-associated gene cassette actually represents a novel fosfomycin resistance determinant. Antimicrob. Agents Chemother. 49:2573. 10.1128/AAC.49.6.2573.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neuner EA, Sekeres J, Hall GS, van Duin D. 2012. Experience with fosfomycin for treatment of urinary tract infections due to multidrug-resistant organisms. Antimicrob. Agents Chemother. 56:5744–5748. 10.1128/AAC.00402-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soraci AL, Perez DS, Martinez G, Dieguez S, Tapia MO, Amanto F, Harkes R, Romano O. 2011. Disodium-fosfomycin pharmacokinetics and bioavailability in post weaning piglets. Res. Vet. Sci. 90:498–502. 10.1016/j.rvsc.2010.07.011 [DOI] [PubMed] [Google Scholar]