FIG 2.
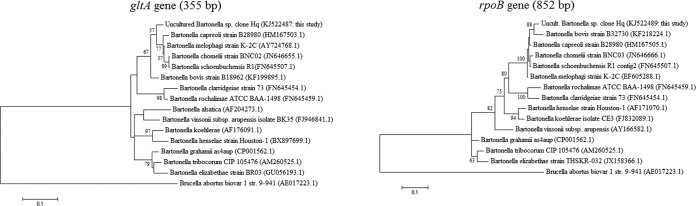
Maximum-likelihood phylogenetic trees based on partial rpoB (825-bp) and gltA (355-bp) genes. Bootstrap values higher than 50% are indicated. Phylogenetic trees were obtained from the uncultured Bartonella sp. clone Hq sequences detected from Haematopinus quadripertusus lice and common Bartonella species sequences, including ruminant-associated Bartonella species (sequences were obtained from the GenBank database, and accession numbers are indicated in parentheses). The Brucella abortus biovar 1, strain 9-941, sequence was used as an outgroup.
