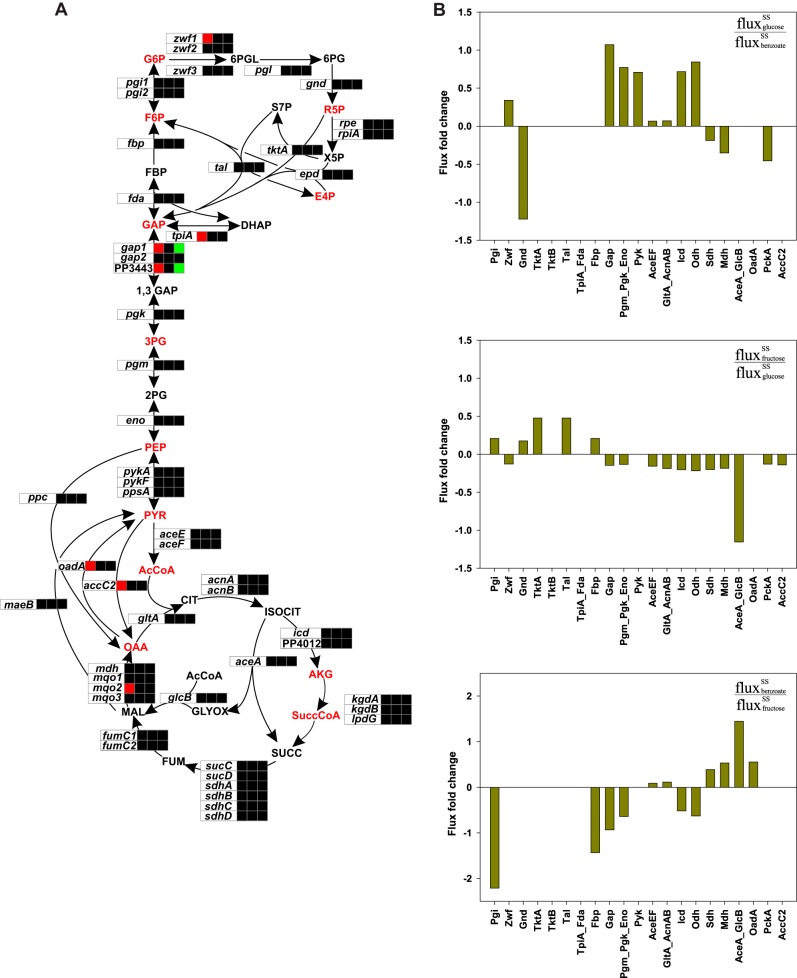
FIG 7.
Transcriptional (A) and flux (B) fold changes of the central metabolic reactions between steady states (SS) on glucose, fructose, and benzoate. (A) The three boxes adjacent to the genes denote the gene expression levels at steady state on glucose (glucose/benzoate transcript ratio), fructose (fructose/glucose transcript ratio), and benzoate (benzoate/fructose transcript ratio), in sequential order. Black boxes indicate no differential regulation. Metabolic intermediates in red type represent the 12 biomass precursor metabolites, which are abbreviated as follows: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; R5P, ribulose-5-phosphate; E4P, erythrose-4-phosphate; GAP, glyceraldehyde-3-phosphate; 3PG, phosphoglyceric acid; PEP, phosphoenolpyruvate; PYR, pyruvate; AcCoA, acetyl coenzyme A; SuccCoA, succinyl coenzyme A; OAA, oxaloacetate; AKG, alpha-ketoglutarate. (B) The fold changes in fluxes were calculated from the net fluxes with the following formula: log10(fluxC source/fluxprevious C source).