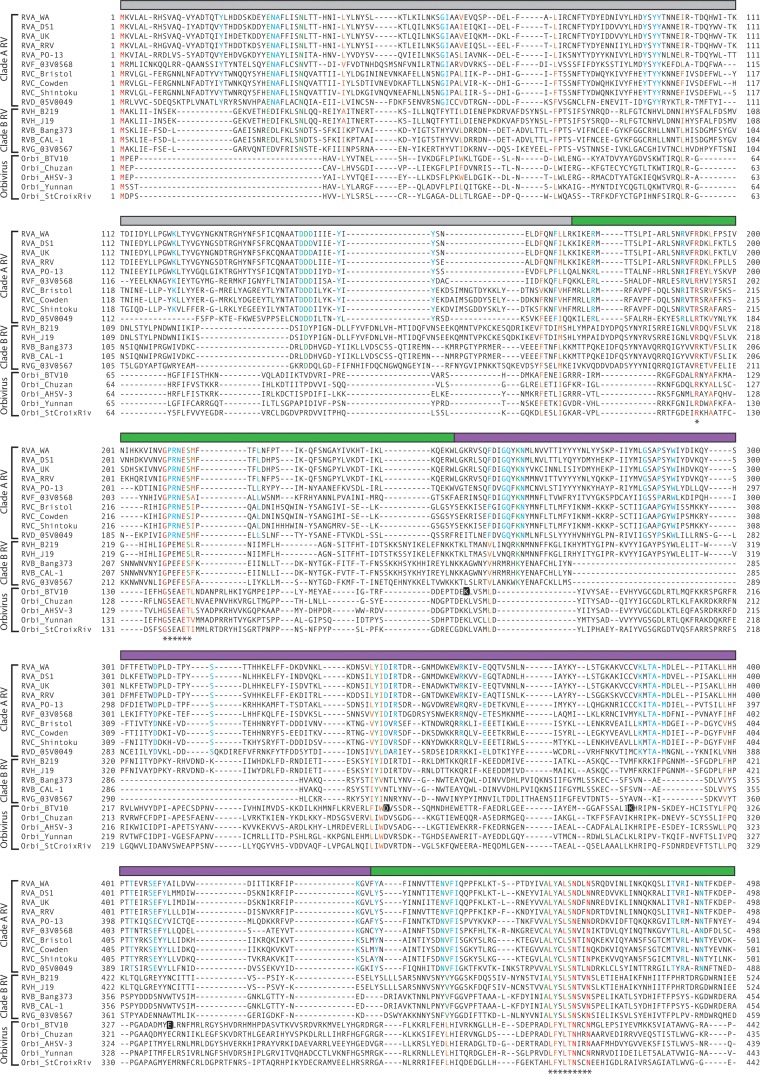
FIG 2.
Sequence conservation among rotavirus and orbivirus capping enzymes. Amino acid alignment of rotavirus (VP3) and orbivirus (VP4) capping enzymes. The rotavirus species, strain name, and clade are indicated; orbivirus (Orbi) strain names are indicated. Aligned residues are colored according to their conservation in the alignment: red, identical for all viruses; orange, similar for all viruses; green, identical for all rotaviruses; and cyan, identical for all clade A rotaviruses. The colored bar above each row of the alignment indicates a structural domain of BTV10 VP4 (gray, N terminus; green, N7-MTase; purple, 2′-O-MTase; blue, GTase/RTPase) or a predicted structural domain of RVA RRV VP3 (cyan, PDE). Asterisks indicate motifs or residues discussed in the text. Black squares highlight residues of the BTV10 VP4 2′-O-MTase KDKE motif. Predicted HΦ(T/S)Φ motifs in the PDE domain of rotavirus VP3 are outlined.